182:
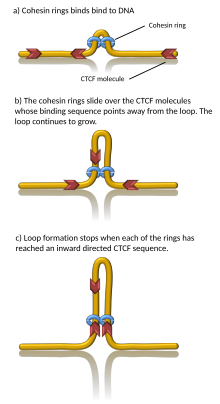
encounters a chromatin-bound CTCF protein, typically located at the boundary of a TAD. In this way, TAD boundaries can be brought together as the anchors of a chromatin loop. Indeed, in vitro, cohesin has been observed to processively extrude DNA loops in an ATP-dependent manner and stall at CTCF. However, some in vitro data indicates that the observed loops may be artifacts. Importantly, since cohesins can dynamically unbind from chromatin, this model suggests that TADs (and associated chromatin loops) are dynamic, transient structures, in agreement with in vivo observations.
31:
268:
155:
181:
Computer simulations have shown that chromatin loop extrusion driven by cohesin motors can generate TADs. In the loop extrusion model, cohesin binds chromatin, pulls it in, and extrudes chromatin to progressively grow a loop. Chromatin on both sides of the cohesin complex is extruded until cohesin
104:
and DNA elements are associated with TAD boundaries. However, the handcuff model and the loop extrusion model describe the TAD formation by the aid of CTCF and cohesin proteins. Furthermore, it has been proposed that the stiffness of TAD boundaries itself could cause the domain insulation and TAD
141:
TAD locations are defined by applying an algorithm to Hi-C data. For example, TADs are often called according to the so-called "directionality index". The directionality index is calculated for individual 40kb bins, by collecting the reads that fall in the bin, and observing whether their paired
258:
For example, genomic structural variants that disrupt TAD boundaries have been reported to cause developmental disorders such as human limb malformations. Additionally, several studies have provided evidence that the disruption or rearrangement of TAD boundaries can provide growth advantages to
142:
reads map upstream or downstream of the bin (read pairs are required to span no more than 2Mb). A positive value indicates that more read pairs lie downstream than upstream, and a negative value indicates the reverse. Mathematically, the directionality index is a signed chi-square statistic.
145:
The development of specialized genome browsers and visualization tools such as
Juicebox, HiGlass/HiPiler, The 3D Genome Browser, 3DIV, 3D-GNOME, and TADKB have enabled us to visualize the TAD organization of regions of interest in different cell types.
230:
Computer simulations have shown that transcription-induced supercoiling of chromatin fibres can explain how TADs are formed and how they can assure very efficient interactions between enhancers and their cognate promoters located in the same TAD.
185:
Other mechanisms for TAD formation have been suggested. For example, some simulations suggest that transcription-generated supercoiling can relocalize cohesin to TAD boundaries or that passively diffusing cohesin “slip links” can generate TADs.
170:. It is also unknown what components are required at TAD boundaries; however, in mammalian cells, it has been shown that these boundary regions have comparatively high levels of CTCF binding. In addition, some types of genes (such as
54:, and they have similar sizes in non-mammalian species. Boundaries at both side of these domains are conserved between different mammalian cell types and even across species and are highly enriched with
1986:
Mach P, Kos PI, Zhan Y, Cramard J, Gaudin S, Tünnermann J, et al. (2022-03-03). "Live-cell imaging and physical modeling reveal control of chromosome folding dynamics by cohesin and CTCF".
85:
interaction to each TAD; however, a recent study uncouples TAD organization and gene expression. Disruption of TAD boundaries are found to be associated with wide range of diseases such as
2620:
Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, et al. (October 2016). "Formation of new chromatin domains determines pathogenicity of genomic duplications".
239:
Replication timing domains have been shown to be associated with TADs as their boundary is co localized with the boundaries of TADs that are located at either sides of compartments.
1965:
Beckwith KS, Ødegård-Fougner Ø, Morero NR, Barton C, Schueder F, Tang W, et al. (2022-05-02). "Visualization of loop extrusion by DNA nanoscale tracing in single human cells".
1760:
Davidson IF, Barth R, Zaczek M, van der Torre J, Tang W, Nagasaka K, et al. (2022-09-09). "CTCF is a DNA-tension-dependent barrier to cohesin-mediated DNA loop extrusion".
199:
TADs have been reported to be relatively constant between different cell types (in stem cells and blood cells, for example), and even between species in specific cases.
2165:
Brackley CA, Johnson J, Michieletto D, Morozov AN, Nicodemi M, Cook PR, Marenduzzo D (September 2017). "Nonequilibrium
Chromosome Looping via Molecular Slip Links".
50:
within a TAD physically interact with each other more frequently than with sequences outside the TAD. The median size of a TAD in mouse cells is 880
1781:
Zhang H, Shi Z, Banigan EJ, Kim Y, Yu H, Bai X, Finkelstein IJ (2022-10-07). "CTCF and R-loops are boundaries of cohesin-mediated DNA looping".
2118:"Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of TADs in interphase chromosomes"
1851:
Ryu JK, Bouchoux C, Liu HW, Kim E, Minamino M, de Groot R, Katan AJ, Bonato A, Marenduzzo D, Michieletto D, Uhlmann F (February 2021).
118:
2331:
Jost D, Vaillant C, Meister P (February 2017). "Coupling 1D modifications and 3D nuclear organization: data, models and function".
281:
Lamina-associated domains (LADs) are parts of the chromatin that heavily interact with the lamina, a network-like structure at the
298:
114:
97:, and F-syndrome, and number of brain disorders like Hypoplastic corpus callosum and Adult-onset demyelinating leukodystrophy.
73:
The functions of TADs are not fully understood and are still a matter of debate. Most of the studies indicate TADs regulate
2507:
Lupiáñez DG, Spielmann M, Mundlos S (April 2016). "Breaking TADs: How
Alterations of Chromatin Domains Result in Disease".
2982:
1268:"The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions"
113:
TADs are defined as regions whose DNA sequences preferentially contact each other. They were discovered in 2012 using
920:
de Laat W, Duboule D (October 2013). "Topology of mammalian developmental enhancers and their regulatory landscapes".
2886:
Li M, Liu GH, Izpisua
Belmonte JC (July 2012). "Navigating the epigenetic landscape of pluripotent stem cells".
2977:
2738:
Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. (January 2016).
219:
to delete the relevant region of the genome) can allow new promoter-enhancer contacts to form. This can affect
1595:
Golfier S, Quail T, Kimura H, Brugués J (May 2020). Dekker, Struhl K, Mirny LA, Musacchio A, Marko JF (eds.).
1485:"Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes"
569:
Krijger PH, de Laat W (December 2016). "Regulation of disease-associated gene expression in the 3D genome".
1415:
317:
240:
100:
The mechanisms underlying TAD formation are also complex and not yet fully elucidated, though a number of
35:
30:
2797:"Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking"
2554:"Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions"
2751:
2694:
2629:
2415:
Marchal C, Sima J, Gilbert DM (December 2019). "Control of DNA replication timing in the 3D genome".
2235:
2184:
2020:
1987:
1966:
1921:
1864:
1782:
1761:
1716:
1659:
1496:
1114:
987:
929:
525:
457:
2602:
1219:"HiPiler: Visual Exploration of Large Genome Interaction Matrices with Interactive Small Multiples"
1166:
Kerpedjiev P, Abdennur N, Lekschas F, McCallum C, Dinkla K, Strobelt H, et al. (August 2018).
1031:
Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, et al. (February 2012).
286:
212:
208:
82:
78:
267:
2911:
2663:
2440:
2208:
2174:
1685:
1577:
953:
850:
594:
367:
342:
Pombo A, Dillon N (April 2015). "Three-dimensional genome architecture: players and mechanisms".
175:
67:
2795:
Weischenfeldt J, Dubash T, Drainas AP, Mardin BR, Chen Y, Stütz AM, et al. (January 2017).
2007:
Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N, et al. (April 2017).
1908:
Gabriele M, Brandão HB, Grosse-Holz S, Jha A, Dailey GM, Cattoglio C, et al. (April 2022).
271:
LADs (dark gray lines) and proteins that interact with them. Lamina is indicated by green curve.
259:
certain cancers, such as T-cell acute lymphoblastic leukemia (T-ALL), gliomas, and lung cancer.
17:
2903:
2868:
2826:
2777:
2720:
2655:
2583:
2534:
2489:
2432:
2397:
2348:
2310:
2261:
2200:
2147:
2098:
2046:
1947:
1890:
1833:
1742:
1677:
1628:
1569:
1524:
1465:
1397:
1348:
1299:
1248:
1199:
1148:
1095:
1054:
1013:
945:
902:
842:
794:
753:
700:
651:
586:
551:
483:
419:
359:
243:, DNA loops formed by CTCF/cohesin-bound regions, are proposed to functionally underlie TADs.
2552:
Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, et al. (May 2015).
2279:
Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S (March 2015).
2226:
Yamamoto T, Schiessel H (September 2017). "Osmotic mechanism of the loop extrusion process".
1483:
Sanborn AL; Rao SS; Huang SC; Durand NC; Huntley MH; Jewett AI; et al. (November 2015).
1366:
Szalaj P, Michalski PJ, Wróblewski P, Tang Z, Kadlof M, Mazzocco G, et al. (July 2016).
622:"Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression"
2895:
2860:
2816:
2808:
2767:
2759:
2710:
2702:
2645:
2637:
2573:
2565:
2524:
2516:
2479:
2471:
2458:
Ji X, Dadon DB, Powell BE, Fan ZP, Borges-Rivera D, Shachar S, et al. (February 2016).
2424:
2387:
2379:
2340:
2300:
2292:
2251:
2243:
2192:
2137:
2129:
2088:
2080:
2036:
2028:
2009:"Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition"
1937:
1929:
1880:
1872:
1823:
1815:
1732:
1724:
1667:
1618:
1608:
1559:
1514:
1504:
1455:
1447:
1387:
1379:
1338:
1330:
1289:
1279:
1238:
1230:
1189:
1179:
1138:
1130:
1085:
1044:
1003:
995:
937:
892:
884:
832:
824:
784:
743:
735:
690:
682:
641:
633:
578:
541:
533:
473:
465:
444:
Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. (April 2012).
409:
401:
351:
282:
2281:"Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture"
1033:"Three-dimensional folding and functional organization principles of the Drosophila genome"
514:"Topological domains in mammalian genomes identified by analysis of chromatin interactions"
2848:
2650:
2529:
815:
Spielmann M, Lupiáñez DG, Mundlos S (July 2018). "Structural variation in the 3D genome".
405:
305:
252:
220:
154:
101:
74:
162:
A number of proteins are known to be associated with TAD formation including the protein
2755:
2698:
2633:
2239:
2188:
2024:
1925:
1910:"Dynamics of CTCF- and cohesin-mediated chromatin looping revealed by live-cell imaging"
1868:
1720:
1663:
1564:
1547:
1500:
991:
933:
529:
461:
2821:
2796:
2772:
2739:
2715:
2682:
2578:
2553:
2484:
2459:
2392:
2367:
2305:
2280:
2142:
2117:
2093:
2068:
2041:
2008:
1942:
1909:
1885:
1852:
1828:
1803:
1737:
1704:
1623:
1596:
1519:
1484:
1460:
1435:
1416:
TADKB: Family classification and a knowledge base of topologically associating domains.
1392:
1367:
1343:
1318:
1294:
1267:
1243:
1218:
1194:
1167:
1143:
1118:
1008:
975:
897:
872:
748:
723:
695:
670:
646:
621:
617:
546:
513:
478:
445:
414:
389:
276:
94:
90:
133:
genomes. In bacteria, they are referred to as
Chromosomal Interacting Domains (CIDs).
2971:
2444:
1689:
1581:
837:
613:
2915:
2212:
1119:"Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom"
854:
598:
2681:
Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, et al. (March 2016).
2667:
2196:
957:
371:
171:
63:
47:
2947:
2296:
1451:
888:
285:. LADs consist mostly of transcriptionally silent chromatin, being enriched with
121:. They have been shown to be present in multiple species, including fruit flies (
1819:
1543:
224:
2569:
2475:
2383:
2247:
1489:
Proceedings of the
National Academy of Sciences of the United States of America
1168:"HiGlass: web-based visual exploration and analysis of genome interaction maps"
1134:
1049:
1032:
446:"Spatial partitioning of the regulatory landscape of the X-inactivation centre"
2864:
2520:
2428:
2344:
1992:
1971:
1787:
1766:
1434:
Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA (May 2016).
1368:"3D-GNOME: an integrated web service for structural modeling of the 3D genome"
1284:
1234:
1184:
828:
739:
719:
686:
637:
290:
122:
1266:
Wang Y, Song F, Zhang B, Zhang L, Xu J, Kuang D, et al. (October 2018).
223:
nearby - such misregulation has been shown to cause limb malformations (e.g.
2706:
1933:
1853:"Bridging-induced phase separation induced by cohesin SMC protein complexes"
1728:
1672:
1647:
1509:
1419:
512:
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. (April 2012).
51:
2907:
2872:
2830:
2781:
2724:
2659:
2587:
2538:
2493:
2436:
2401:
2352:
2314:
2265:
2204:
2151:
2102:
2050:
1951:
1894:
1876:
1837:
1746:
1681:
1632:
1573:
1528:
1469:
1401:
1352:
1317:
Yang D, Jang I, Choi J, Kim MS, Lee AJ, Kim H, et al. (January 2018).
1303:
1252:
1203:
1152:
1099:
1058:
1017:
999:
949:
906:
846:
798:
757:
704:
655:
590:
555:
487:
423:
363:
2133:
2084:
1646:
Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters JM (December 2019).
1597:"Cohesin and condensin extrude DNA loops in a cell cycle-dependent manner"
1334:
1217:
Lekschas F, Bach B, Kerpedjiev P, Gehlenborg N, Pfister H (January 2018).
789:
772:
178:) appear near TAD boundaries more often than would be expected by chance.
70:) appear near TAD boundaries more often than would be expected by chance.
2851:(August 2016). "On TADs and LADs: Spatial Control Over Gene Expression".
2683:"Activation of proto-oncogenes by disruption of chromosome neighborhoods"
1383:
582:
294:
215:
do not cross TAD boundaries. Removing a TAD boundary (for example, using
2763:
2641:
2032:
1613:
941:
537:
469:
2368:"Comparing 3D Genome Organization in Multiple Species Using Phylo-HMRF"
1090:
1073:
301:
167:
59:
2256:
2740:"Insulator dysfunction and oncogene activation in IDH mutant gliomas"
1074:"Visualising three-dimensional genome organisation in two dimensions"
976:"Principles of genome folding into topologically associating domains"
216:
86:
2899:
2812:
1113:
Durand NC; Robinson JT; Shamim MS; Machol I; Mesirov JP; Lander ES;
355:
2179:
266:
153:
130:
126:
2962:
2957:
2932:
163:
55:
2460:"3D Chromosome Regulatory Landscape of Human Pluripotent Cells"
1804:"DNA sliding and loop formation by E. coli SMC complex: MukBEF"
2942:
1703:
Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H (December 2019).
2116:
Racko D, Benedetti F, Dorier J, Stasiak A (February 2018).
2603:"A Family's Shared Defect Sheds Light on the Human Genome"
2067:
Racko D, Benedetti F, Dorier J, Stasiak A (January 2019).
2937:
390:"The Three-Dimensional Organization of Mammalian Genomes"
235:
Relationship with other structural features of the genome
46:(TAD) is a self-interacting genomic region, meaning that
2952:
1223:
873:"Chromatin Domains: The Unit of Chromosome Organization"
2366:
Yang Y, Zhang Y, Ren B, Dixon JR, Ma J (June 2019).
1436:"Formation of Chromosomal Domains by Loop Extrusion"
2842:
2840:
1319:"3DIV: A 3D-genome Interaction Viewer and database"
308:. LADs have CTCF-binding sites at their periphery.
1548:"Organization of Chromosomal DNA by SMC Complexes"
612:Ghavi-Helm Y; Jankowski A; Meiers S; Viales RR;
1072:Ing-Simmons E, Vaquerizas JM (September 2019).
974:Szabo Q, Bantignies F, Cavalli G (April 2019).
394:Annual Review of Cell and Developmental Biology
1705:"Human cohesin compacts DNA by loop extrusion"
773:"Disruption of the 3D cancer genome blueprint"
207:The majority of observed interactions between
2062:
2060:
969:
967:
771:Achinger-Kawecka J, Clark SJ (January 2017).
728:Current Opinion in Genetics & Development
675:Current Opinion in Genetics & Development
507:
505:
503:
501:
499:
497:
255:of nearby genes, and this can cause disease.
8:
439:
437:
435:
433:
337:
335:
333:
251:Disruption of TAD boundaries can affect the
203:Relationship with promoter-enhancer contacts
62:. In addition, some types of genes (such as
2326:
2324:
2820:
2771:
2714:
2649:
2577:
2528:
2483:
2391:
2304:
2255:
2178:
2141:
2092:
2040:
1991:
1970:
1941:
1884:
1827:
1786:
1765:
1736:
1671:
1622:
1612:
1563:
1518:
1508:
1459:
1420:https://doi.org/10.1186/s12864-019-5551-2
1391:
1342:
1293:
1283:
1242:
1193:
1183:
1142:
1089:
1048:
1007:
896:
836:
788:
747:
694:
645:
545:
477:
413:
34:Topologically associating domains within
871:Dixon JR, Gorkin DU, Ren B (June 2016).
158:DNA loop extrusion through cohesin rings
89:, variety of limb malformations such as
29:
329:
2888:Nature Reviews. Molecular Cell Biology
2417:Nature Reviews. Molecular Cell Biology
669:Corces MR, Corces VG (February 2016).
571:Nature Reviews. Molecular Cell Biology
344:Nature Reviews. Molecular Cell Biology
1648:"DNA loop extrusion by human cohesin"
1429:
1427:
1414:Liu, T., Porter, J., Zhao, C. et al.
866:
864:
810:
808:
671:"The three-dimensional cancer genome"
406:10.1146/annurev-cellbio-100616-060531
7:
724:"TAD disruption as oncogenic driver"
383:
381:
1808:Biochemistry and Biophysics Reports
1565:10.1146/annurev-genet-112618-043633
25:
44:topologically associating domain
38:, their borders and interactions
18:Topologically Associating Domain
2333:Current Opinion in Cell Biology
115:chromosome conformation capture
27:Self-interacting genomic region
2651:11858/00-001M-0000-002C-010A-3
2530:11858/00-001M-0000-002E-1D1D-D
2197:10.1103/PhysRevLett.119.138101
137:Analytical tools and databases
1:
1418:BMC Genomics 20, 217 (2019).
283:inner membrane of the nucleus
2297:10.1016/j.celrep.2015.02.004
1802:Man, Zhou (September 2022).
1452:10.1016/j.celrep.2016.04.085
889:10.1016/j.molcel.2016.05.018
388:Yu M, Ren B (October 2017).
1820:10.1016/j.bbrep.2022.101297
56:CCCTC-binding factor (CTCF)
2999:
2570:10.1016/j.cell.2015.04.004
2476:10.1016/j.stem.2015.11.007
2384:10.1016/j.cels.2019.05.011
2248:10.1103/PhysRevE.96.030402
1135:10.1016/j.cels.2015.07.012
1050:10.1016/j.cell.2012.01.010
274:
2865:10.1016/j.tig.2016.05.004
2521:10.1016/j.tig.2016.01.003
2429:10.1038/s41580-019-0162-y
2345:10.1016/j.ceb.2016.12.001
1993:10.1101/2022.03.03.482826
1972:10.1101/2021.04.12.439407
1788:10.1101/2022.09.15.508177
1767:10.1101/2022.09.08.507093
1552:Annual Review of Genetics
1285:10.1186/s13059-018-1519-9
1235:10.1109/TVCG.2017.2745978
1185:10.1186/s13059-018-1486-1
838:21.11116/0000-0003-610A-5
829:10.1038/s41576-018-0007-0
740:10.1016/j.gde.2016.03.008
687:10.1016/j.gde.2016.01.002
638:10.1038/s41588-019-0462-3
263:Lamina-associated domains
1542:Yatskevich S; Rhodes J;
817:Nature Reviews. Genetics
166:and the protein complex
2707:10.1126/science.aad9024
2601:Angier N (2017-01-09).
2167:Physical Review Letters
2069:"Are TADs supercoiled?"
1934:10.1126/science.abn6583
1729:10.1126/science.aaz4475
1673:10.1126/science.aaz3418
1510:10.1073/pnas.1518552112
241:Insulated neighborhoods
150:Mechanisms of formation
109:Discovery and diversity
2122:Nucleic Acids Research
2073:Nucleic Acids Research
1877:10.1126/sciadv.abe5905
1372:Nucleic Acids Research
1323:Nucleic Acids Research
1000:10.1126/sciadv.aaw1668
318:Insulated neighborhood
272:
227:) in humans and mice.
159:
39:
36:chromosome territories
2847:Gonzalez-Sandoval A;
790:10.2217/epi-2016-0111
297:); which is a common
270:
157:
117:techniques including
33:
2983:Nuclear organization
583:10.1038/nrm.2016.138
129:, plants, fungi and
2764:10.1038/nature16490
2756:2016Natur.529..110F
2699:2016Sci...351.1454H
2693:(6280): 1454–1458.
2642:10.1038/nature19800
2634:2016Natur.538..265F
2240:2017PhRvE..96c0402Y
2189:2017PhRvL.119m8101B
2134:10.1093/nar/gkx1123
2085:10.1093/nar/gky1091
2033:10.1038/nature21711
2025:2017Natur.544..110F
1926:2022Sci...376..496G
1869:2021SciA....7.5905R
1721:2019Sci...366.1345K
1715:(6471): 1345–1349.
1664:2019Sci...366.1338D
1658:(6471): 1338–1345.
1614:10.7554/eLife.53885
1501:2015PNAS..112E6456S
1495:(47): E6456–E6465.
1335:10.1093/nar/gkx1017
992:2019SciA....5.1668S
942:10.1038/nature12753
934:2013Natur.502..499D
538:10.1038/nature11082
530:2012Natur.485..376D
470:10.1038/nature11049
462:2012Natur.485..381N
2853:Trends in Genetics
2607:The New York Times
2509:Trends in Genetics
2378:(6): 494–505.e14.
1384:10.1093/nar/gkw437
1091:10.1242/dev.177162
273:
176:housekeeping genes
160:
68:housekeeping genes
40:
2948:3D Genome Browser
2750:(7584): 110–114.
2628:(7624): 265–269.
2228:Physical Review E
2019:(7648): 110–114.
1920:(6592): 496–501.
1546:(December 2019).
1378:(W1): W288–W293.
928:(7472): 499–506.
722:(February 2016).
524:(7398): 376–380.
456:(7398): 381–385.
299:posttranslational
102:protein complexes
16:(Redirected from
2990:
2920:
2919:
2883:
2877:
2876:
2844:
2835:
2834:
2824:
2792:
2786:
2785:
2775:
2735:
2729:
2728:
2718:
2678:
2672:
2671:
2653:
2617:
2611:
2610:
2598:
2592:
2591:
2581:
2564:(5): 1012–1025.
2549:
2543:
2542:
2532:
2504:
2498:
2497:
2487:
2455:
2449:
2448:
2412:
2406:
2405:
2395:
2363:
2357:
2356:
2328:
2319:
2318:
2308:
2291:(8): 1297–1309.
2276:
2270:
2269:
2259:
2223:
2217:
2216:
2182:
2162:
2156:
2155:
2145:
2128:(4): 1648–1660.
2113:
2107:
2106:
2096:
2064:
2055:
2054:
2044:
2004:
1998:
1997:
1995:
1983:
1977:
1976:
1974:
1962:
1956:
1955:
1945:
1905:
1899:
1898:
1888:
1857:Science Advances
1848:
1842:
1841:
1831:
1799:
1793:
1792:
1790:
1778:
1772:
1771:
1769:
1757:
1751:
1750:
1740:
1700:
1694:
1693:
1675:
1643:
1637:
1636:
1626:
1616:
1592:
1586:
1585:
1567:
1539:
1533:
1532:
1522:
1512:
1480:
1474:
1473:
1463:
1446:(9): 2038–2049.
1431:
1422:
1412:
1406:
1405:
1395:
1363:
1357:
1356:
1346:
1314:
1308:
1307:
1297:
1287:
1263:
1257:
1256:
1246:
1214:
1208:
1207:
1197:
1187:
1163:
1157:
1156:
1146:
1110:
1104:
1103:
1093:
1069:
1063:
1062:
1052:
1028:
1022:
1021:
1011:
980:Science Advances
971:
962:
961:
917:
911:
910:
900:
868:
859:
858:
840:
812:
803:
802:
792:
768:
762:
761:
751:
715:
709:
708:
698:
666:
660:
659:
649:
632:(8): 1272–1282.
609:
603:
602:
566:
560:
559:
549:
509:
492:
491:
481:
441:
428:
427:
417:
385:
376:
375:
339:
304:modification of
77:by limiting the
21:
2998:
2997:
2993:
2992:
2991:
2989:
2988:
2987:
2978:Gene expression
2968:
2967:
2929:
2924:
2923:
2900:10.1038/nrm3393
2885:
2884:
2880:
2846:
2845:
2838:
2813:10.1038/ng.3722
2801:Nature Genetics
2794:
2793:
2789:
2737:
2736:
2732:
2680:
2679:
2675:
2619:
2618:
2614:
2600:
2599:
2595:
2551:
2550:
2546:
2506:
2505:
2501:
2457:
2456:
2452:
2423:(12): 721–737.
2414:
2413:
2409:
2365:
2364:
2360:
2330:
2329:
2322:
2278:
2277:
2273:
2234:(3–1): 030402.
2225:
2224:
2220:
2164:
2163:
2159:
2115:
2114:
2110:
2066:
2065:
2058:
2006:
2005:
2001:
1985:
1984:
1980:
1964:
1963:
1959:
1907:
1906:
1902:
1863:(7): eabe5905.
1850:
1849:
1845:
1801:
1800:
1796:
1780:
1779:
1775:
1759:
1758:
1754:
1702:
1701:
1697:
1645:
1644:
1640:
1594:
1593:
1589:
1541:
1540:
1536:
1482:
1481:
1477:
1433:
1432:
1425:
1413:
1409:
1365:
1364:
1360:
1329:(D1): D52–D57.
1316:
1315:
1311:
1265:
1264:
1260:
1216:
1215:
1211:
1165:
1164:
1160:
1112:
1111:
1107:
1071:
1070:
1066:
1030:
1029:
1025:
986:(4): eaaw1668.
973:
972:
965:
919:
918:
914:
870:
869:
862:
814:
813:
806:
770:
769:
765:
717:
716:
712:
668:
667:
663:
626:Nature Genetics
620:(August 2019).
611:
610:
606:
577:(12): 771–782.
568:
567:
563:
511:
510:
495:
443:
442:
431:
387:
386:
379:
356:10.1038/nrm3965
341:
340:
331:
326:
314:
306:heterochromatin
279:
265:
249:
247:Role in disease
237:
221:gene expression
205:
197:
192:
152:
139:
111:
75:gene expression
28:
23:
22:
15:
12:
11:
5:
2996:
2994:
2986:
2985:
2980:
2970:
2969:
2966:
2965:
2960:
2955:
2950:
2945:
2940:
2935:
2928:
2927:External links
2925:
2922:
2921:
2894:(8): 524–535.
2878:
2859:(8): 485–495.
2836:
2787:
2730:
2673:
2612:
2593:
2544:
2515:(4): 225–237.
2499:
2470:(2): 262–275.
2464:Cell Stem Cell
2450:
2407:
2358:
2320:
2271:
2218:
2173:(13): 138101.
2157:
2108:
2079:(2): 521–532.
2056:
1999:
1978:
1957:
1900:
1843:
1794:
1773:
1752:
1695:
1638:
1587:
1558:(1): 445–482.
1534:
1475:
1423:
1407:
1358:
1309:
1272:Genome Biology
1258:
1229:(1): 522–531.
1209:
1172:Genome Biology
1158:
1105:
1084:(19): 99–101.
1064:
1043:(3): 458–472.
1023:
963:
912:
883:(5): 668–680.
877:Molecular Cell
860:
823:(7): 453–467.
804:
763:
710:
661:
604:
561:
493:
429:
377:
350:(4): 245–257.
328:
327:
325:
322:
321:
320:
313:
310:
277:Nuclear lamina
275:Main article:
264:
261:
248:
245:
236:
233:
204:
201:
196:
193:
191:
188:
151:
148:
138:
135:
110:
107:
95:Cooks syndrome
91:synpolydactyly
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
2995:
2984:
2981:
2979:
2976:
2975:
2973:
2964:
2961:
2959:
2956:
2954:
2951:
2949:
2946:
2944:
2941:
2939:
2936:
2934:
2931:
2930:
2926:
2917:
2913:
2909:
2905:
2901:
2897:
2893:
2889:
2882:
2879:
2874:
2870:
2866:
2862:
2858:
2854:
2850:
2843:
2841:
2837:
2832:
2828:
2823:
2818:
2814:
2810:
2806:
2802:
2798:
2791:
2788:
2783:
2779:
2774:
2769:
2765:
2761:
2757:
2753:
2749:
2745:
2741:
2734:
2731:
2726:
2722:
2717:
2712:
2708:
2704:
2700:
2696:
2692:
2688:
2684:
2677:
2674:
2669:
2665:
2661:
2657:
2652:
2647:
2643:
2639:
2635:
2631:
2627:
2623:
2616:
2613:
2608:
2604:
2597:
2594:
2589:
2585:
2580:
2575:
2571:
2567:
2563:
2559:
2555:
2548:
2545:
2540:
2536:
2531:
2526:
2522:
2518:
2514:
2510:
2503:
2500:
2495:
2491:
2486:
2481:
2477:
2473:
2469:
2465:
2461:
2454:
2451:
2446:
2442:
2438:
2434:
2430:
2426:
2422:
2418:
2411:
2408:
2403:
2399:
2394:
2389:
2385:
2381:
2377:
2373:
2369:
2362:
2359:
2354:
2350:
2346:
2342:
2338:
2334:
2327:
2325:
2321:
2316:
2312:
2307:
2302:
2298:
2294:
2290:
2286:
2282:
2275:
2272:
2267:
2263:
2258:
2253:
2249:
2245:
2241:
2237:
2233:
2229:
2222:
2219:
2214:
2210:
2206:
2202:
2198:
2194:
2190:
2186:
2181:
2176:
2172:
2168:
2161:
2158:
2153:
2149:
2144:
2139:
2135:
2131:
2127:
2123:
2119:
2112:
2109:
2104:
2100:
2095:
2090:
2086:
2082:
2078:
2074:
2070:
2063:
2061:
2057:
2052:
2048:
2043:
2038:
2034:
2030:
2026:
2022:
2018:
2014:
2010:
2003:
2000:
1994:
1989:
1982:
1979:
1973:
1968:
1961:
1958:
1953:
1949:
1944:
1939:
1935:
1931:
1927:
1923:
1919:
1915:
1911:
1904:
1901:
1896:
1892:
1887:
1882:
1878:
1874:
1870:
1866:
1862:
1858:
1854:
1847:
1844:
1839:
1835:
1830:
1825:
1821:
1817:
1813:
1809:
1805:
1798:
1795:
1789:
1784:
1777:
1774:
1768:
1763:
1756:
1753:
1748:
1744:
1739:
1734:
1730:
1726:
1722:
1718:
1714:
1710:
1706:
1699:
1696:
1691:
1687:
1683:
1679:
1674:
1669:
1665:
1661:
1657:
1653:
1649:
1642:
1639:
1634:
1630:
1625:
1620:
1615:
1610:
1606:
1602:
1598:
1591:
1588:
1583:
1579:
1575:
1571:
1566:
1561:
1557:
1553:
1549:
1545:
1538:
1535:
1530:
1526:
1521:
1516:
1511:
1506:
1502:
1498:
1494:
1490:
1486:
1479:
1476:
1471:
1467:
1462:
1457:
1453:
1449:
1445:
1441:
1437:
1430:
1428:
1424:
1421:
1417:
1411:
1408:
1403:
1399:
1394:
1389:
1385:
1381:
1377:
1373:
1369:
1362:
1359:
1354:
1350:
1345:
1340:
1336:
1332:
1328:
1324:
1320:
1313:
1310:
1305:
1301:
1296:
1291:
1286:
1281:
1277:
1273:
1269:
1262:
1259:
1254:
1250:
1245:
1240:
1236:
1232:
1228:
1224:
1220:
1213:
1210:
1205:
1201:
1196:
1191:
1186:
1181:
1177:
1173:
1169:
1162:
1159:
1154:
1150:
1145:
1140:
1136:
1132:
1129:(1): 99–101.
1128:
1124:
1120:
1117:(July 2016).
1116:
1109:
1106:
1101:
1097:
1092:
1087:
1083:
1079:
1075:
1068:
1065:
1060:
1056:
1051:
1046:
1042:
1038:
1034:
1027:
1024:
1019:
1015:
1010:
1005:
1001:
997:
993:
989:
985:
981:
977:
970:
968:
964:
959:
955:
951:
947:
943:
939:
935:
931:
927:
923:
916:
913:
908:
904:
899:
894:
890:
886:
882:
878:
874:
867:
865:
861:
856:
852:
848:
844:
839:
834:
830:
826:
822:
818:
811:
809:
805:
800:
796:
791:
786:
782:
778:
774:
767:
764:
759:
755:
750:
745:
741:
737:
733:
729:
725:
721:
714:
711:
706:
702:
697:
692:
688:
684:
680:
676:
672:
665:
662:
657:
653:
648:
643:
639:
635:
631:
627:
623:
619:
615:
608:
605:
600:
596:
592:
588:
584:
580:
576:
572:
565:
562:
557:
553:
548:
543:
539:
535:
531:
527:
523:
519:
515:
508:
506:
504:
502:
500:
498:
494:
489:
485:
480:
475:
471:
467:
463:
459:
455:
451:
447:
440:
438:
436:
434:
430:
425:
421:
416:
411:
407:
403:
399:
395:
391:
384:
382:
378:
373:
369:
365:
361:
357:
353:
349:
345:
338:
336:
334:
330:
323:
319:
316:
315:
311:
309:
307:
303:
300:
296:
292:
288:
287:trimethylated
284:
278:
269:
262:
260:
256:
254:
246:
244:
242:
234:
232:
228:
226:
222:
218:
214:
210:
202:
200:
194:
189:
187:
183:
179:
177:
173:
169:
165:
156:
149:
147:
143:
136:
134:
132:
128:
124:
120:
116:
108:
106:
103:
98:
96:
92:
88:
84:
80:
76:
71:
69:
65:
61:
57:
53:
49:
48:DNA sequences
45:
37:
32:
19:
2891:
2887:
2881:
2856:
2852:
2807:(1): 65–74.
2804:
2800:
2790:
2747:
2743:
2733:
2690:
2686:
2676:
2625:
2621:
2615:
2606:
2596:
2561:
2557:
2547:
2512:
2508:
2502:
2467:
2463:
2453:
2420:
2416:
2410:
2375:
2372:Cell Systems
2371:
2361:
2336:
2332:
2288:
2285:Cell Reports
2284:
2274:
2231:
2227:
2221:
2170:
2166:
2160:
2125:
2121:
2111:
2076:
2072:
2016:
2012:
2002:
1981:
1960:
1917:
1913:
1903:
1860:
1856:
1846:
1811:
1807:
1797:
1776:
1755:
1712:
1708:
1698:
1655:
1651:
1641:
1604:
1600:
1590:
1555:
1551:
1537:
1492:
1488:
1478:
1443:
1440:Cell Reports
1439:
1410:
1375:
1371:
1361:
1326:
1322:
1312:
1275:
1271:
1261:
1226:
1222:
1212:
1175:
1171:
1161:
1126:
1123:Cell Systems
1122:
1108:
1081:
1077:
1067:
1040:
1036:
1026:
983:
979:
925:
921:
915:
880:
876:
820:
816:
783:(1): 47–55.
780:
776:
766:
731:
727:
713:
678:
674:
664:
629:
625:
607:
574:
570:
564:
521:
517:
453:
449:
397:
393:
347:
343:
280:
257:
250:
238:
229:
206:
198:
195:Conservation
184:
180:
172:transfer RNA
161:
144:
140:
112:
99:
72:
64:transfer RNA
43:
41:
1078:Development
777:Epigenomics
718:Valton AL;
400:: 265–289.
225:polydactyly
105:formation.
2972:Categories
2257:1887/58394
2180:1612.07256
1814:: 101297.
1607:: e53885.
1278:(1): 151.
1178:(1): 125.
618:Furlong EE
324:References
291:histone H3
253:expression
190:Properties
174:genes and
123:Drosophila
66:genes and
2849:Gasser SM
2445:201714312
2339:: 20–27.
1690:208228309
1582:203653572
1544:Nasmyth K
734:: 34–40.
614:Korbel JO
289:Lys27 on
213:enhancers
209:promoters
2958:3D-GNOME
2933:Juicebox
2916:22524502
2908:22820889
2873:27312344
2831:27869826
2782:26700815
2725:26940867
2660:27706140
2588:25959774
2539:26862051
2494:26686465
2437:31477886
2402:31229558
2353:28040646
2315:25732821
2266:29346962
2213:14706723
2205:29341686
2152:29140466
2103:30395328
2051:28355183
1952:35420890
1895:33568486
1838:35770038
1747:31780627
1682:31753851
1633:32396063
1574:31577909
1529:26499245
1470:27210764
1402:27185892
1353:29106613
1304:30286773
1253:28866592
1204:30143029
1153:27467250
1115:Aiden EL
1100:31558569
1059:22265598
1018:30989119
950:24153303
907:27259200
855:22325904
847:29692413
799:27936932
758:27111891
720:Dekker J
705:26855137
656:31308546
599:11484886
591:27826147
556:22495300
488:22495304
424:28783961
364:25757416
312:See also
295:H3K27me3
293:, (i.e.
83:promoter
79:enhancer
2943:HiPiler
2938:HiGlass
2822:5791882
2773:4831574
2752:Bibcode
2716:4884612
2695:Bibcode
2687:Science
2668:4463482
2630:Bibcode
2579:4791538
2485:4848748
2393:6706282
2306:4542312
2236:Bibcode
2185:Bibcode
2143:5829651
2094:6344874
2042:5639698
2021:Bibcode
1988:bioRxiv
1967:bioRxiv
1943:9069445
1922:Bibcode
1914:Science
1886:7875533
1865:Bibcode
1829:9234588
1783:bioRxiv
1762:bioRxiv
1738:7387118
1717:Bibcode
1709:Science
1660:Bibcode
1652:Science
1624:7316503
1520:4664323
1497:Bibcode
1461:4889513
1393:4987952
1344:5753379
1295:6172833
1244:6038708
1195:6109259
1144:5596920
1009:6457944
988:Bibcode
958:4468533
930:Bibcode
898:5371509
749:4880504
696:4880523
681:: 1–7.
647:7116017
547:3356448
526:Bibcode
479:3555144
458:Bibcode
415:5837811
372:6713103
302:histone
168:cohesin
60:cohesin
2914:
2906:
2871:
2829:
2819:
2780:
2770:
2744:Nature
2723:
2713:
2666:
2658:
2622:Nature
2586:
2576:
2537:
2492:
2482:
2443:
2435:
2400:
2390:
2351:
2313:
2303:
2264:
2211:
2203:
2150:
2140:
2101:
2091:
2049:
2039:
2013:Nature
1990:
1969:
1950:
1940:
1893:
1883:
1836:
1826:
1785:
1764:
1745:
1735:
1688:
1680:
1631:
1621:
1580:
1572:
1527:
1517:
1468:
1458:
1400:
1390:
1351:
1341:
1302:
1292:
1251:
1241:
1202:
1192:
1151:
1141:
1098:
1057:
1016:
1006:
956:
948:
922:Nature
905:
895:
853:
845:
797:
756:
746:
703:
693:
654:
644:
597:
589:
554:
544:
518:Nature
486:
476:
450:Nature
422:
412:
370:
362:
217:CRISPR
87:cancer
2963:TADKB
2912:S2CID
2664:S2CID
2441:S2CID
2209:S2CID
2175:arXiv
1686:S2CID
1601:eLife
1578:S2CID
954:S2CID
851:S2CID
595:S2CID
368:S2CID
131:human
127:mouse
2953:3DIV
2904:PMID
2869:PMID
2827:PMID
2778:PMID
2721:PMID
2656:PMID
2584:PMID
2558:Cell
2535:PMID
2490:PMID
2433:PMID
2398:PMID
2349:PMID
2311:PMID
2262:PMID
2201:PMID
2148:PMID
2099:PMID
2047:PMID
1948:PMID
1891:PMID
1834:PMID
1743:PMID
1678:PMID
1629:PMID
1570:PMID
1525:PMID
1466:PMID
1398:PMID
1349:PMID
1300:PMID
1249:PMID
1200:PMID
1149:PMID
1096:PMID
1055:PMID
1037:Cell
1014:PMID
946:PMID
903:PMID
843:PMID
795:PMID
754:PMID
701:PMID
652:PMID
587:PMID
552:PMID
484:PMID
420:PMID
360:PMID
211:and
164:CTCF
119:Hi-C
58:and
2896:doi
2861:doi
2817:PMC
2809:doi
2768:PMC
2760:doi
2748:529
2711:PMC
2703:doi
2691:351
2646:hdl
2638:doi
2626:538
2574:PMC
2566:doi
2562:161
2525:hdl
2517:doi
2480:PMC
2472:doi
2425:doi
2388:PMC
2380:doi
2341:doi
2301:PMC
2293:doi
2252:hdl
2244:doi
2193:doi
2171:119
2138:PMC
2130:doi
2089:PMC
2081:doi
2037:PMC
2029:doi
2017:544
1938:PMC
1930:doi
1918:376
1881:PMC
1873:doi
1824:PMC
1816:doi
1733:PMC
1725:doi
1713:366
1668:doi
1656:366
1619:PMC
1609:doi
1560:doi
1515:PMC
1505:doi
1493:112
1456:PMC
1448:doi
1388:PMC
1380:doi
1339:PMC
1331:doi
1290:PMC
1280:doi
1239:PMC
1231:doi
1190:PMC
1180:doi
1139:PMC
1131:doi
1086:doi
1082:146
1045:doi
1041:148
1004:PMC
996:doi
938:doi
926:502
893:PMC
885:doi
833:hdl
825:doi
785:doi
744:PMC
736:doi
691:PMC
683:doi
642:PMC
634:doi
579:doi
542:PMC
534:doi
522:485
474:PMC
466:doi
454:485
410:PMC
402:doi
352:doi
125:),
2974::
2910:.
2902:.
2892:13
2890:.
2867:.
2857:32
2855:.
2839:^
2825:.
2815:.
2805:49
2803:.
2799:.
2776:.
2766:.
2758:.
2746:.
2742:.
2719:.
2709:.
2701:.
2689:.
2685:.
2662:.
2654:.
2644:.
2636:.
2624:.
2605:.
2582:.
2572:.
2560:.
2556:.
2533:.
2523:.
2513:32
2511:.
2488:.
2478:.
2468:18
2466:.
2462:.
2439:.
2431:.
2421:20
2419:.
2396:.
2386:.
2374:.
2370:.
2347:.
2337:44
2335:.
2323:^
2309:.
2299:.
2289:10
2287:.
2283:.
2260:.
2250:.
2242:.
2232:96
2230:.
2207:.
2199:.
2191:.
2183:.
2169:.
2146:.
2136:.
2126:46
2124:.
2120:.
2097:.
2087:.
2077:47
2075:.
2071:.
2059:^
2045:.
2035:.
2027:.
2015:.
2011:.
1946:.
1936:.
1928:.
1916:.
1912:.
1889:.
1879:.
1871:.
1859:.
1855:.
1832:.
1822:.
1812:31
1810:.
1806:.
1741:.
1731:.
1723:.
1711:.
1707:.
1684:.
1676:.
1666:.
1654:.
1650:.
1627:.
1617:.
1603:.
1599:.
1576:.
1568:.
1556:53
1554:.
1550:.
1523:.
1513:.
1503:.
1491:.
1487:.
1464:.
1454:.
1444:15
1442:.
1438:.
1426:^
1396:.
1386:.
1376:44
1374:.
1370:.
1347:.
1337:.
1327:46
1325:.
1321:.
1298:.
1288:.
1276:19
1274:.
1270:.
1247:.
1237:.
1227:24
1225:.
1221:.
1198:.
1188:.
1176:19
1174:.
1170:.
1147:.
1137:.
1125:.
1121:.
1094:.
1080:.
1076:.
1053:.
1039:.
1035:.
1012:.
1002:.
994:.
982:.
978:.
966:^
952:.
944:.
936:.
924:.
901:.
891:.
881:62
879:.
875:.
863:^
849:.
841:.
831:.
821:19
819:.
807:^
793:.
779:.
775:.
752:.
742:.
732:36
730:.
726:.
699:.
689:.
679:36
677:.
673:.
650:.
640:.
630:51
628:.
624:.
616:;
593:.
585:.
575:17
573:.
550:.
540:.
532:.
520:.
516:.
496:^
482:.
472:.
464:.
452:.
448:.
432:^
418:.
408:.
398:33
396:.
392:.
380:^
366:.
358:.
348:16
346:.
332:^
93:,
52:kb
42:A
2918:.
2898::
2875:.
2863::
2833:.
2811::
2784:.
2762::
2754::
2727:.
2705::
2697::
2670:.
2648::
2640::
2632::
2609:.
2590:.
2568::
2541:.
2527::
2519::
2496:.
2474::
2447:.
2427::
2404:.
2382::
2376:8
2355:.
2343::
2317:.
2295::
2268:.
2254::
2246::
2238::
2215:.
2195::
2187::
2177::
2154:.
2132::
2105:.
2083::
2053:.
2031::
2023::
1996:.
1975:.
1954:.
1932::
1924::
1897:.
1875::
1867::
1861:7
1840:.
1818::
1791:.
1770:.
1749:.
1727::
1719::
1692:.
1670::
1662::
1635:.
1611::
1605:9
1584:.
1562::
1531:.
1507::
1499::
1472:.
1450::
1404:.
1382::
1355:.
1333::
1306:.
1282::
1255:.
1233::
1206:.
1182::
1155:.
1133::
1127:3
1102:.
1088::
1061:.
1047::
1020:.
998::
990::
984:5
960:.
940::
932::
909:.
887::
857:.
835::
827::
801:.
787::
781:9
760:.
738::
707:.
685::
658:.
636::
601:.
581::
558:.
536::
528::
490:.
468::
460::
426:.
404::
374:.
354::
81:-
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.