29:
1958:
935:"Cytosolic mercaptopyruvate sulfurtransferase is evolutionarily related to mitochondrial rhodanese. Striking similarity in active site amino acid sequence and the increase in the mercaptopyruvate sulfurtransferase activity of rhodanese by site-directed mutagenesis"
1099:
Billaut-Laden I, Rat E, Allorge D, Crunelle-Thibaut A, Cauffiez C, Chevalier D, Lo-Guidice JM, Broly F (August 2006). "Evidence for a functional genetic polymorphism of the human mercaptopyruvate sulfurtransferase (MPST), a cyanide detoxification enzyme".
1162:
Nagahara N, Ito T, Kitamura H, Nishino T (September 1998). "Tissue and subcellular distribution of mercaptopyruvate sulfurtransferase in the rat: confocal laser fluorescence and immunoelectron microscopic studies combined with biochemical analysis".
1271:
Gai JW, Qin W, Liu M, Wang HF, Zhang M, Li M, Zhou WH, Ma QT, Liu GM, Song WH, Jin J, Ma HS (April 2016). "Expression profile of hydrogen sulfide and its synthases correlates with tumor stage and grade in urothelial cell carcinoma of bladder".
548:
The biological function of MPST remains unclear. It may be involved in cyanide detoxification, biosynthesis of thiosulfate, production of the signalling molecule hydrogen sulfide, or the degradation of cysteine.
1496:
482:
of this enzyme class is 3-mercaptopyruvate:cyanide sulfurtransferase. This enzyme is also called beta-mercaptopyruvate sulfurtransferase and in the older literature, human liver rhodanese.
1489:
977:
Shibuya N, Mikami Y, Kimura Y, Nagahara N, Kimura H (November 2009). "Vascular endothelium expresses 3-mercaptopyruvate sulfurtransferase and produces hydrogen sulfide".
204:
710:), a rare inheritable disorder associated with oversecretion of mercaptolactate-cysteine disulfide in urine. Fewer than 1,000 people have this rare disorder in the
517:
The encoded cytoplasmic protein is a member of the rhodanese family but is not rhodanese itself, which is found only in mitochondria. MPST protein consists of 317
1482:
277:
223:
1561:
1551:
1978:
738:
samples. Protein levels and catalytic activities of MPST increased with the increase of malignant degrees in human bladder tissues and human
1644:
1988:
301:
575:
The persulfide group is labile, and can transfer S to other groups, such as cyanide. It is also susceptible to reduction to release
1677:
1223:
815:
289:
1050:
Kuo MM, Kim DH, Jandu S, Bergman Y, Tan S, Wang H, Pandey DR, Abraham TP, Shoukas AA, Berkowitz DE, Santhanam L (January 2016).
216:
1598:
282:
1135:
Ubuka T, Hosaki Y, Nishina H, Ikeda T (1985). "3-Mercaptopyruvate sulfurtransferase activity in guinea pig and rat tissues".
143:
1833:
1410:
Vachek H, Wood JL (January 1972). "Purification and properties of mercaptopyruvate sulfur transferase of
Escherichia coli".
1247:
1012:
Vachek H, Wood JL (January 1972). "Purification and properties of mercaptopyruvate sulfur transferase of
Escherichia coli".
167:
1534:
832:
Patterson SE, Moeller B, Nagasawa HT, Vince R, Crankshaw DL, Briggs J, Stutelberg MW, Vinnakota CV, Logue BA (June 2016).
739:
1948:
1625:
28:
1818:
1934:
1921:
1908:
1895:
1882:
1869:
1856:
1635:
1613:
1581:
1522:
1828:
1782:
1725:
1593:
1513:
1248:"Beta-mercaptolactate cysteine disulfiduria - About the Disease - Genetic and Rare Diseases Information Center"
426:
354:
161:
54:
1052:"MPST but not CSE is the primary regulator of hydrogen sulfide production and function in the coronary artery"
675:
587:
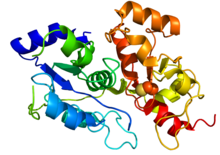
43:. Rainbow colored (N-terminus blue and C-terminus red). The catalytic cysteine is depicted by spheres.
1730:
891:"Cloning and sequence analysis of the human liver rhodanese: comparison with the bovine and chicken enzymes"
148:
742:(UCB) cell lines and this may promote the application of MPST novel enzymes to UCB diagnosis or treatment.
610:
S. MPST catalyzes the transfer of a sulfur atom from mercaptopyruvate to sulfur acceptors like cyanides or
1556:
294:
1751:
1670:
1603:
595:
371:
228:
1823:
136:
1571:
845:
71:
1787:
529:
518:
453:
66:
164:
1720:
1474:
1381:
Sorbo B (May 1957). "Enzymic transfer of sulfur from mercaptopyruvate to sulfate or sulfinates".
1188:
603:
445:
88:
650:
599:
552:
MPST reacts with 3-MP to convert a cysteinyl residue to the persulfide-containing intermediate:
1983:
1526:
1462:
1427:
1398:
1369:
1334:
1289:
1229:
1219:
1180:
1144:
1117:
1081:
1029:
994:
956:
912:
871:
794:
763:"Structure and kinetic analysis of H2S production by human mercaptopyruvate sulfurtransferase"
614:
compounds. Thus it is also considered to participate in cysteine degradation. MPST generates H
467:
441:
313:
155:
35:
509:. Alternatively spliced transcript variants encoding the same protein have been identified.
1766:
1761:
1735:
1663:
1585:
1452:
1419:
1390:
1359:
1324:
1281:
1211:
1172:
1109:
1071:
1063:
1021:
986:
946:
902:
861:
853:
784:
774:
654:
646:
576:
540:
rhodanese domain. The protein can function as a monomer or as a disulfide-linked homodimer.
366:
375:
1813:
1797:
1710:
1566:
683:
619:
479:
395:
359:
124:
849:
638:
MPST is expressed in a number of tissues, including kidney, liver, lung, heart, muscle,
100:
1962:
1851:
1792:
1076:
1051:
866:
833:
789:
762:
735:
715:
525:
199:
59:
1457:
1440:
1364:
1347:
1329:
1312:
907:
890:
179:
1972:
1756:
1715:
1423:
1394:
1215:
1025:
731:
711:
174:
1192:
1705:
687:
623:
1313:"Specificity studies on the beta-mercaptopyruvate-cyanide transsulfuration system"
1285:
1113:
330:
1929:
1864:
1700:
1505:
1067:
719:
449:
405:
183:
1957:
1617:
537:
502:
418:
337:
1206:
Sörbo B (1987). "3-Mercaptopyruvate, 3-mercaptolactate and mercaptoacetate".
951:
934:
1903:
1877:
1539:
779:
522:
470:, especially since it occurs widely in the cytosol and distributed broadly.
437:
33:
Crystallographic structure of 3-mercaptopyruvate sulfurtransferase based on
1402:
1373:
1338:
1293:
1121:
1085:
998:
875:
798:
1466:
1431:
1233:
1184:
1176:
1148:
1033:
960:
916:
533:
342:
1441:"A study of the cooper content of beta-mercaptopyruvate trans-sulfurase"
990:
857:
429:
131:
112:
318:
39:
1916:
1686:
1509:
639:
433:
325:
211:
107:
95:
83:
452:, specifically the sulfurtransferases. This enzyme participates in
1890:
611:
506:
1056:
American
Journal of Physiology. Heart and Circulatory Physiology
761:
Yadav PK, Yamada K, Chiku T, Koutmos M, Banerjee R (July 2013).
497:
460:
306:
119:
1659:
1478:
889:
Pallini R, Guazzi GC, Cannella C, Cacace MG (October 1991).
690:, namely vasodilation. This has important implications for H
626:, namely vasodilation. This has important implications for H
466:
The enzyme is of interest because it provides a pathway for
1348:"Enzymatic formation of polysulfides from mercaptopyruvate"
694:
S-based therapy in healthy and diseased coronary arteries.
630:
S-based therapy in healthy and diseased coronary arteries.
1655:
1210:. Methods in Enzymology. Vol. 143. pp. 178–82.
722:, hypolasia of the ear cartilage and umbilical hernia.
1439:
Van den Hamer CJ, Morell AG, Scheinberg IH (May 1967).
834:"Development of sulfanegen for mass cyanide casualties"
714:. Symptoms and signs include: high forehead, seizures,
532:
suggesting a common evolutionary origin. The catalytic
816:"Entrez Gene: MPST mercaptopyruvate sulfurtransferase"
1946:
686:, mediating its effects through direct modulation of
657:. Thus MPST may participate in cysteine degradation.
622:, mediating its effects through direct modulation of
594:
S by MPST is a two-step reaction. In the first step,
730:
Immunoreactivity for MPST was detected in malignant
590:(CSE) and MPST. l-cysteine-dependent production of H
1842:
1806:
1775:
1744:
1693:
1634:
1612:
1580:
1521:
1137:
Physiological
Chemistry and Physics and Medical NMR
895:
401:
391:
386:
365:
353:
348:
336:
324:
312:
300:
288:
276:
268:
260:
252:
247:
242:
222:
210:
198:
193:
173:
154:
142:
130:
118:
106:
94:
82:
77:
65:
53:
48:
21:
1412:Biochimica et Biophysica Acta (BBA) - Enzymology
1014:Biochimica et Biophysica Acta (BBA) - Enzymology
653:, is derived from cysteine by the action of an
606:(3-MP). Subsequently, MPST converts 3-MP into H
972:
970:
933:Nagahara N, Okazaki T, Nishino T (July 1995).
1671:
1490:
702:Deficiency in MPST has been found related to
8:
1045:
1043:
928:
926:
1678:
1664:
1656:
1497:
1483:
1475:
838:Annals of the New York Academy of Sciences
383:
190:
27:
1456:
1363:
1328:
1304:Further reading (mainly older literature)
1075:
950:
906:
865:
788:
778:
810:
808:
1953:
750:
674:S is produced from MPST (as well as by
448:. This enzyme belongs to the family of
756:
754:
521:and weighs 35250Da. MPST contains two
505:location of 22q12.3 and consists of 6
239:
18:
1562:thiosulfate—dithiol sulfurtransferase
1311:Fiedler H, Wood JL (September 1956).
704:mercaptolactate-cysteine disulfiduria
698:Mercaptolactate-cysteine disulfiduria
7:
1645:Coenzyme-B sulfoethylthiotransferase
1547:3-mercaptopyruvate sulfurtransferase
827:
825:
740:urothelial cell carcinoma of bladder
423:3-mercaptopyruvate sulfurtransferase
243:3-mercaptopyruvate sulfurtransferase
22:3-mercaptopyruvate sulfurtransferase
1552:thiosulfate—thiol sulfurtransferase
1445:The Journal of Biological Chemistry
1352:The Journal of Biological Chemistry
1317:The Journal of Biological Chemistry
939:The Journal of Biological Chemistry
767:The Journal of Biological Chemistry
14:
1346:Hylin JW, Wood JL (August 1959).
586:S is produced from l-cysteine by
1956:
1599:Tyrosylprotein sulfotransferase
1165:Histochemistry and Cell Biology
598:converts l-cysteine along with
1:
1535:Thiosulfate sulfurtransferase
1458:10.1016/S0021-9258(18)95992-2
1383:Biochimica et Biophysica Acta
1365:10.1016/S0021-9258(18)69881-3
1330:10.1016/S0021-9258(19)50803-1
1286:10.1016/j.urolonc.2015.06.020
1208:Sulfur and Sulfur Amino Acids
908:10.1016/S0006-291X(05)81148-9
1979:Genes on human chromosome 22
1424:10.1016/0005-2744(72)90973-4
1395:10.1016/0006-3002(57)90201-9
1216:10.1016/0076-6879(87)43033-4
1114:10.1016/j.toxlet.2006.02.002
1026:10.1016/0005-2744(72)90973-4
1068:10.1152/ajpheart.00574.2014
666:Hydrogen sulfide production
536:residue only exists in the
2005:
1989:Enzymes of known structure
1626:Propionate CoA-transferase
1834:Michaelis–Menten kinetics
1252:rarediseases.info.nih.gov
468:detoxification of cyanide
382:
189:
26:
1726:Diffusion-limited enzyme
1594:Alcohol sulfotransferase
952:10.1074/jbc.270.27.16230
979:Journal of Biochemistry
780:10.1074/jbc.M113.466177
634:Biological significance
456:. It is encoded by the
1557:tRNA sulfurtransferase
734:and muscular layer of
544:Function and mechanism
1819:Eadie–Hofstee diagram
1752:Allosteric regulation
1604:Aryl sulfotransferase
1177:10.1007/s004180050286
676:cystathionine-γ-lyase
596:cysteine transaminase
588:cystathionine-γ-lyase
272:human liver rhodanese
1829:Lineweaver–Burk plot
1572:cysteine desulfurase
678:). MPST generates H
530:secondary structures
1512:-containing group (
850:2016NYASA1374..202P
649:(3-MP), along with
519:amino acid residues
454:cysteine metabolism
1788:Enzyme superfamily
1721:Enzyme promiscuity
1527:Sulfurtransferases
1102:Toxicology Letters
858:10.1111/nyas.13114
604:3-mercaptopyruvate
446:3-mercaptopyruvate
442:chemical reactions
1944:
1943:
1653:
1652:
1586:Sulfotransferases
1280:(4): 166.e15–20.
1274:Urologic Oncology
991:10.1093/jb/mvp111
501:gene lies on the
415:
414:
411:
410:
238:
237:
234:
233:
137:metabolic pathway
1996:
1961:
1960:
1952:
1824:Hanes–Woolf plot
1767:Enzyme activator
1762:Enzyme inhibitor
1736:Enzyme catalysis
1680:
1673:
1666:
1657:
1618:CoA-transferases
1499:
1492:
1485:
1476:
1470:
1460:
1435:
1406:
1377:
1367:
1342:
1332:
1298:
1297:
1268:
1262:
1261:
1259:
1258:
1244:
1238:
1237:
1203:
1197:
1196:
1159:
1153:
1152:
1132:
1126:
1125:
1096:
1090:
1089:
1079:
1047:
1038:
1037:
1009:
1003:
1002:
974:
965:
964:
954:
930:
921:
920:
910:
886:
880:
879:
869:
829:
820:
819:
812:
803:
802:
792:
782:
773:(27): 20002–13.
758:
655:aminotransferase
647:Mercaptopyruvate
577:hydrogen sulfide
384:
240:
191:
42:
31:
19:
16:Class of enzymes
2004:
2003:
1999:
1998:
1997:
1995:
1994:
1993:
1969:
1968:
1967:
1955:
1947:
1945:
1940:
1852:Oxidoreductases
1838:
1814:Enzyme kinetics
1802:
1798:List of enzymes
1771:
1740:
1711:Catalytic triad
1689:
1684:
1654:
1649:
1630:
1608:
1576:
1567:biotin synthase
1517:
1503:
1473:
1438:
1409:
1380:
1345:
1310:
1306:
1301:
1270:
1269:
1265:
1256:
1254:
1246:
1245:
1241:
1226:
1205:
1204:
1200:
1161:
1160:
1156:
1134:
1133:
1129:
1098:
1097:
1093:
1049:
1048:
1041:
1011:
1010:
1006:
976:
975:
968:
945:(27): 16230–5.
932:
931:
924:
888:
887:
883:
831:
830:
823:
814:
813:
806:
760:
759:
752:
748:
728:
708:Ampola syndrome
700:
693:
684:coronary artery
681:
673:
668:
663:
651:α-ketoglutarate
636:
629:
620:coronary artery
617:
609:
600:α-ketoglutarate
593:
585:
571:
567:
564:→ RSSH + CH
563:
559:
546:
515:
493:
488:
480:systematic name
476:
264:MST, TST2, TUM1
44:
34:
17:
12:
11:
5:
2002:
2000:
1992:
1991:
1986:
1981:
1971:
1970:
1966:
1965:
1942:
1941:
1939:
1938:
1925:
1912:
1899:
1886:
1873:
1860:
1846:
1844:
1840:
1839:
1837:
1836:
1831:
1826:
1821:
1816:
1810:
1808:
1804:
1803:
1801:
1800:
1795:
1790:
1785:
1779:
1777:
1776:Classification
1773:
1772:
1770:
1769:
1764:
1759:
1754:
1748:
1746:
1742:
1741:
1739:
1738:
1733:
1728:
1723:
1718:
1713:
1708:
1703:
1697:
1695:
1691:
1690:
1685:
1683:
1682:
1675:
1668:
1660:
1651:
1650:
1648:
1647:
1641:
1639:
1632:
1631:
1629:
1628:
1622:
1620:
1610:
1609:
1607:
1606:
1601:
1596:
1590:
1588:
1578:
1577:
1575:
1574:
1569:
1564:
1559:
1554:
1549:
1544:
1543:
1542:
1531:
1529:
1519:
1518:
1504:
1502:
1501:
1494:
1487:
1479:
1472:
1471:
1451:(10): 2514–6.
1436:
1407:
1378:
1343:
1307:
1305:
1302:
1300:
1299:
1263:
1239:
1224:
1198:
1154:
1127:
1091:
1039:
1004:
966:
922:
881:
821:
804:
749:
747:
744:
736:bladder cancer
727:
726:Bladder cancer
724:
716:arachnodactyly
699:
696:
691:
679:
671:
667:
664:
662:
659:
635:
632:
627:
615:
607:
591:
583:
573:
572:
569:
565:
561:
557:
545:
542:
514:
511:
492:
489:
487:
484:
475:
472:
413:
412:
409:
408:
403:
399:
398:
393:
389:
388:
380:
379:
369:
363:
362:
357:
351:
350:
346:
345:
340:
334:
333:
328:
322:
321:
316:
310:
309:
304:
298:
297:
292:
286:
285:
280:
274:
273:
270:
266:
265:
262:
258:
257:
254:
250:
249:
245:
244:
236:
235:
232:
231:
226:
220:
219:
214:
208:
207:
202:
196:
195:
187:
186:
177:
171:
170:
159:
152:
151:
146:
140:
139:
134:
128:
127:
122:
116:
115:
110:
104:
103:
98:
92:
91:
86:
80:
79:
75:
74:
69:
63:
62:
57:
51:
50:
46:
45:
32:
24:
23:
15:
13:
10:
9:
6:
4:
3:
2:
2001:
1990:
1987:
1985:
1982:
1980:
1977:
1976:
1974:
1964:
1959:
1954:
1950:
1936:
1932:
1931:
1926:
1923:
1919:
1918:
1913:
1910:
1906:
1905:
1900:
1897:
1893:
1892:
1887:
1884:
1880:
1879:
1874:
1871:
1867:
1866:
1861:
1858:
1854:
1853:
1848:
1847:
1845:
1841:
1835:
1832:
1830:
1827:
1825:
1822:
1820:
1817:
1815:
1812:
1811:
1809:
1805:
1799:
1796:
1794:
1793:Enzyme family
1791:
1789:
1786:
1784:
1781:
1780:
1778:
1774:
1768:
1765:
1763:
1760:
1758:
1757:Cooperativity
1755:
1753:
1750:
1749:
1747:
1743:
1737:
1734:
1732:
1729:
1727:
1724:
1722:
1719:
1717:
1716:Oxyanion hole
1714:
1712:
1709:
1707:
1704:
1702:
1699:
1698:
1696:
1692:
1688:
1681:
1676:
1674:
1669:
1667:
1662:
1661:
1658:
1646:
1643:
1642:
1640:
1637:
1633:
1627:
1624:
1623:
1621:
1619:
1615:
1611:
1605:
1602:
1600:
1597:
1595:
1592:
1591:
1589:
1587:
1583:
1579:
1573:
1570:
1568:
1565:
1563:
1560:
1558:
1555:
1553:
1550:
1548:
1545:
1541:
1538:
1537:
1536:
1533:
1532:
1530:
1528:
1524:
1520:
1515:
1511:
1507:
1500:
1495:
1493:
1488:
1486:
1481:
1480:
1477:
1468:
1464:
1459:
1454:
1450:
1446:
1442:
1437:
1433:
1429:
1425:
1421:
1418:(1): 133–46.
1417:
1413:
1408:
1404:
1400:
1396:
1392:
1388:
1384:
1379:
1375:
1371:
1366:
1361:
1358:(8): 2141–4.
1357:
1353:
1349:
1344:
1340:
1336:
1331:
1326:
1323:(1): 387–97.
1322:
1318:
1314:
1309:
1308:
1303:
1295:
1291:
1287:
1283:
1279:
1275:
1267:
1264:
1253:
1249:
1243:
1240:
1235:
1231:
1227:
1225:9780121820435
1221:
1217:
1213:
1209:
1202:
1199:
1194:
1190:
1186:
1182:
1178:
1174:
1171:(3): 243–50.
1170:
1166:
1158:
1155:
1150:
1146:
1142:
1138:
1131:
1128:
1123:
1119:
1115:
1111:
1108:(2): 101–11.
1107:
1103:
1095:
1092:
1087:
1083:
1078:
1073:
1069:
1065:
1061:
1057:
1053:
1046:
1044:
1040:
1035:
1031:
1027:
1023:
1020:(1): 133–46.
1019:
1015:
1008:
1005:
1000:
996:
992:
988:
984:
980:
973:
971:
967:
962:
958:
953:
948:
944:
940:
936:
929:
927:
923:
918:
914:
909:
904:
901:(2): 887–93.
900:
896:
892:
885:
882:
877:
873:
868:
863:
859:
855:
851:
847:
843:
839:
835:
828:
826:
822:
817:
811:
809:
805:
800:
796:
791:
786:
781:
776:
772:
768:
764:
757:
755:
751:
745:
743:
741:
737:
733:
732:uroepithelium
725:
723:
721:
717:
713:
712:United States
709:
705:
697:
695:
689:
685:
677:
665:
660:
658:
656:
652:
648:
643:
642:, and brain.
641:
633:
631:
625:
621:
613:
605:
601:
597:
589:
580:
578:
555:
554:
553:
550:
543:
541:
539:
535:
531:
528:with similar
527:
524:
520:
512:
510:
508:
504:
500:
499:
490:
485:
483:
481:
473:
471:
469:
464:
462:
459:
455:
451:
447:
443:
439:
435:
431:
428:
424:
420:
407:
404:
400:
397:
394:
390:
385:
381:
378:
377:
373:
370:
368:
364:
361:
358:
356:
352:
347:
344:
341:
339:
335:
332:
329:
327:
323:
320:
317:
315:
311:
308:
305:
303:
299:
296:
293:
291:
287:
284:
281:
279:
275:
271:
267:
263:
259:
255:
251:
246:
241:
230:
227:
225:
221:
218:
215:
213:
209:
206:
203:
201:
197:
192:
188:
185:
181:
178:
176:
175:Gene Ontology
172:
169:
166:
163:
160:
157:
153:
150:
147:
145:
141:
138:
135:
133:
129:
126:
123:
121:
117:
114:
113:NiceZyme view
111:
109:
105:
102:
99:
97:
93:
90:
87:
85:
81:
76:
73:
70:
68:
64:
61:
58:
56:
52:
47:
41:
37:
30:
25:
20:
1930:Translocases
1927:
1914:
1901:
1888:
1875:
1865:Transferases
1862:
1849:
1706:Binding site
1546:
1506:Transferases
1448:
1444:
1415:
1411:
1389:(2): 324–9.
1386:
1382:
1355:
1351:
1320:
1316:
1277:
1273:
1266:
1255:. Retrieved
1251:
1242:
1207:
1201:
1168:
1164:
1157:
1140:
1136:
1130:
1105:
1101:
1094:
1062:(1): H71–9.
1059:
1055:
1017:
1013:
1007:
985:(5): 623–6.
982:
978:
942:
938:
898:
894:
884:
844:(1): 202–9.
841:
837:
770:
766:
729:
707:
703:
701:
669:
644:
637:
581:
574:
556:RSH + HSCH
551:
547:
516:
496:
494:
477:
474:Nomenclature
465:
457:
450:transferases
422:
416:
374:
261:Alt. symbols
101:BRENDA entry
1701:Active site
1638:: Alkylthio
1143:(1): 41–3.
720:genu valgum
396:Swiss-model
248:Identifiers
89:IntEnz view
49:Identifiers
1973:Categories
1904:Isomerases
1878:Hydrolases
1745:Regulation
1257:2024-03-09
746:References
661:Biomedical
538:C-terminal
503:chromosome
419:enzymology
392:Structures
387:Search for
349:Other data
269:Alt. names
158:structures
125:KEGG entry
72:9026-05-5
1783:EC number
1540:Rhodanese
523:rhodanese
486:Structure
438:catalyzes
355:EC number
331:NP_066949
278:NCBI gene
78:Databases
1984:EC 2.8.1
1807:Kinetics
1731:Cofactor
1694:Activity
1403:13436433
1374:13673028
1339:13367011
1294:26847849
1193:25430792
1122:16545926
1086:26519030
999:19605461
876:27308865
799:23698001
534:cysteine
432:) is an
406:InterPro
229:proteins
217:articles
205:articles
162:RCSB PDB
1963:Biology
1917:Ligases
1687:Enzymes
1467:6026243
1432:4550801
1234:3657534
1185:9749958
1149:3862140
1077:4796461
1034:4550801
961:7608189
917:1953758
867:4940216
846:Bibcode
790:3707699
526:domains
513:Protein
430:2.8.1.2
402:Domains
372:Chr. 22
360:2.8.1.2
338:UniProt
184:QuickGO
149:profile
132:MetaCyc
67:CAS no.
60:2.8.1.2
1949:Portal
1891:Lyases
1510:sulfur
1465:
1430:
1401:
1372:
1337:
1292:
1232:
1222:
1191:
1183:
1147:
1120:
1084:
1074:
1032:
997:
959:
915:
874:
864:
797:
787:
640:spleen
568:C(O)CO
560:C(O)CO
434:enzyme
343:P25325
326:RefSeq
307:602496
253:Symbol
212:PubMed
194:Search
180:AmiGO
168:PDBsum
108:ExPASy
96:BRENDA
84:IntEnz
55:EC no.
1843:Types
1636:2.8.4
1614:2.8.3
1582:2.8.2
1523:2.8.1
1189:S2CID
682:S in
618:S in
612:thiol
602:into
507:exons
436:that
367:Locus
144:PRIAM
1935:list
1928:EC7
1922:list
1915:EC6
1909:list
1902:EC5
1896:list
1889:EC4
1883:list
1876:EC3
1870:list
1863:EC2
1857:list
1850:EC1
1516:2.8)
1463:PMID
1428:PMID
1399:PMID
1370:PMID
1335:PMID
1290:PMID
1230:PMID
1220:ISBN
1181:PMID
1145:PMID
1118:PMID
1082:PMID
1030:PMID
995:PMID
957:PMID
913:PMID
872:PMID
842:1374
795:PMID
706:(or
498:MPST
495:The
491:Gene
478:The
461:gene
458:MPST
440:the
421:, a
376:q123
319:3OLH
302:OMIM
295:7223
290:HGNC
283:4357
256:MPST
224:NCBI
165:PDBe
120:KEGG
40:3OLH
1453:doi
1449:242
1420:doi
1416:258
1391:doi
1360:doi
1356:234
1325:doi
1321:222
1282:doi
1212:doi
1173:doi
1169:110
1110:doi
1106:165
1072:PMC
1064:doi
1060:310
1022:doi
1018:258
987:doi
983:146
947:doi
943:270
903:doi
899:180
862:PMC
854:doi
785:PMC
775:doi
771:288
444:of
417:In
314:PDB
200:PMC
156:PDB
36:PDB
1975::
1616::
1584::
1525::
1514:EC
1508::
1461:.
1447:.
1443:.
1426:.
1414:.
1397:.
1387:24
1385:.
1368:.
1354:.
1350:.
1333:.
1319:.
1315:.
1288:.
1278:34
1276:.
1250:.
1228:.
1218:.
1187:.
1179:.
1167:.
1141:17
1139:.
1116:.
1104:.
1080:.
1070:.
1058:.
1054:.
1042:^
1028:.
1016:.
993:.
981:.
969:^
955:.
941:.
937:.
925:^
911:.
897:.
893:.
870:.
860:.
852:.
840:.
836:.
824:^
807:^
793:.
783:.
769:.
765:.
753:^
718:,
688:NO
645:3-
624:NO
579:.
463:.
427:EC
182:/
38::
1951::
1937:)
1933:(
1924:)
1920:(
1911:)
1907:(
1898:)
1894:(
1885:)
1881:(
1872:)
1868:(
1859:)
1855:(
1679:e
1672:t
1665:v
1498:e
1491:t
1484:v
1469:.
1455::
1434:.
1422::
1405:.
1393::
1376:.
1362::
1341:.
1327::
1296:.
1284::
1260:.
1236:.
1214::
1195:.
1175::
1151:.
1124:.
1112::
1088:.
1066::
1036:.
1024::
1001:.
989::
963:.
949::
919:.
905::
878:.
856::
848::
818:.
801:.
777::
692:2
680:2
672:2
670:H
628:2
616:2
608:2
592:2
584:2
582:H
570:2
566:3
562:2
558:2
425:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.