81:) are added to single stranded DNA to be sequenced and the incorporation of nucleotide is followed by measuring the light emitted. The intensity of the light determines if 0, 1 or more nucleotides have been incorporated, thus showing how many complementary nucleotides are present on the template strand. The nucleotide mixture is removed before the next nucleotide mixture is added. This process is repeated with each of the four nucleotides until the DNA sequence of the single stranded template is determined.
152:
237:
in the presence of adenosine 5´ phosphosulfate. This ATP acts as a substrate for the luciferase-mediated conversion of luciferin to oxyluciferin that generates visible light in amounts that are proportional to the amount. The light produced in the luciferase-catalyzed reaction is detected by a camera
178:
are sequentially added and removed from the reaction. Light is produced only when the nucleotide solution complements the first unpaired base of the template. The sequence of solutions which produce chemiluminescent signals allows the determination of the sequence of the template.
112:
to be added at the start and kept throughout the procedure, thus providing a simple set-up suitable for automation. An automated instrument based on this principle was introduced to the market the following year by the company
Pyrosequencing.
226:) (dATPαS, which is not a substrate for a luciferase, is added instead of dATP to avoid noise) initiates the second step. DNA polymerase incorporates the correct, complementary dNTPs onto the template. This incorporation releases
124:. This alternative approach for pyrosequencing was based on the original principle of attaching the DNA to be sequenced to a solid support and they showed that sequencing could be performed in a highly parallel manner using a
159:"Sequencing by synthesis" involves taking a single strand of the DNA to be sequenced and then synthesizing its complementary strand enzymatically. The pyrosequencing method is based on detecting the activity of
174:
by synthesizing the complementary strand along it, one base pair at a time, and detecting which base was actually added at each step. The template DNA is immobile, and solutions of A, C, G, and T
131:. This allowed for high-throughput DNA sequencing and an automated instrument was introduced to the market. This became the first next generation sequencing instrument starting a new era in
299:
Currently, a limitation of the method is that the lengths of individual reads of DNA sequence are in the neighborhood of 300-500 nucleotides, shorter than the 800-1000 obtainable with
381:
Nyren, Pettersson and Uhlen (1993) “Solid Phase DNA Minisequencing by an
Enzymatic Luminometric Inorganic Pyrophosphate Detection Assay” Analytical Biochemistry 208 (1), 171-175,
335:
in order to commercialize machinery and reagents for sequencing short stretches of DNA using the pyrosequencing technique. Pyrosequencing AB was listed on the
405:
Nyren and Lundin (1985) “Enzymatic method for continuous monitoring of inorganic pyrophosphate synthesis” Analytiocal
Biochemistry 151 (2): 504-509.
526:
57:
coated magnetic beads with recombinant DNA polymerase lacking 3´to 5´exonuclease activity (proof-reading) and luminescence detection using the
30:
in DNA) based on the "sequencing by synthesis" principle, in which the sequencing is performed by detecting the nucleotide incorporated by a
479:
100:
is introduced to remove nucleotides that are not incorporated by the DNA polymerase. This enabled the enzyme mixture including the
602:
597:
89:
207:
219:
70:
464:
Marguiles et al (2005) “Genome sequencing in microfabricated high-density picolitre reactors” Nature 437, 376-380.
607:
187:
336:
140:
78:
592:
234:
50:
447:
419:
571:
439:
362:
344:
164:
121:
117:
339:
in 1999. It was renamed to
Biotage in 2003. The pyrosequencing business line was acquired by
561:
431:
125:
42:
328:
324:
304:
195:
151:
85:
347:. 454 developed an array-based pyrosequencing technology which emerged as a platform for
93:
46:
352:
348:
308:
300:
191:
160:
136:
101:
66:
31:
23:
586:
227:
35:
451:
356:
116:
A third microfluidic variant of the pyrosequencing method was described in 2005 by
54:
41:
The principle of pyrosequencing was first described in 1993 by, Bertil
Pettersson,
435:
175:
27:
406:
199:
128:
105:
74:
58:
332:
211:
575:
84:
A second solution-based method for pyrosequencing was described in 1998 by
501:
465:
443:
34:. Pyrosequencing relies on light detection based on a chain reaction when
382:
182:
For the solution-based version of pyrosequencing, the single-strand DNA (
132:
307:
more difficult, particularly for sequences containing a large amount of
566:
549:
242:
203:
109:
97:
527:"Roche to close 454 Life Sciences as it reduces gene sequencing focus"
365:
announced the discontinuation of the 454 sequencing platform in 2013.
340:
167:
62:
394:
183:
311:. Lack of proof-reading activity limits accuracy of this method.
303:
methods (e.g. Sanger sequencing). This can make the process of
223:
170:. Essentially, the method allows sequencing a single strand of
393:
Uhlen (1989) ”Magnetic separation of DNA” Nature 340: 733-4,
171:
343:
in 2008. Pyrosequencing technology was further licensed to
418:
Ronaghi, Mostafa; Uhlén, Mathias; Nyrén, Pål (1998-07-17).
249:
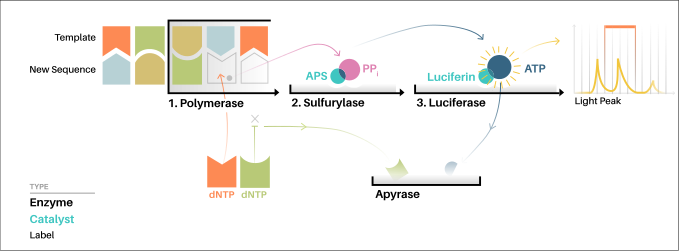
The process can be represented by the following equations:
253:
PPi + APS → ATP + Sulfate (catalyzed by ATP-sulfurylase);
245:, and the reaction can restart with another nucleotide.
241:
Unincorporated nucleotides and ATP are degraded by the
420:"A Sequencing Method Based on Real-Time Pyrophosphate"
256:
ATP + luciferin + O2 → AMP + PPi + oxyluciferin + CO
96:. In this alternative method, an additional enzyme
480:"Pyrosequencing Technology and Platform Overview"
38:is released. Hence, the name pyrosequencing.
8:
407:https://doi.org/10.1016/0003-2697(85)90211-8
135:research, with rapidly falling prices for
565:
550:"Emerging Technologies in DNA Sequencing"
186:) template is hybridized to a sequencing
163:(a DNA synthesizing enzyme) with another
155:The chart shows how pyrosequencing works.
150:
466:https://doi.org/doi:10.1038/nature03959
374:
383:https://doi.org/10.1006/abio.1993.1024
7:
525:Hollmer, Mark (October 17, 2013).
271:APS is adenosine 5-phosphosulfate;
14:
395:https://doi.org/10.1038/340733a0
233:ATP sulfurylase converts PPi to
218:The addition of one of the four
280:AMP is adenosine monophosphate;
260:+ hv (catalyzed by luciferase);
190:and incubated with the enzymes
274:ATP is adenosine triphosphate;
120:and co-workers at the company
1:
220:deoxynucleotide triphosphates
436:10.1126/science.281.5375.363
61:enzyme. A mixture of three
208:adenosine 5´ phosphosulfate
624:
349:large-scale DNA sequencing
238:and analyzed in a program.
206:, and with the substrates
26:(determining the order of
337:Stockholm Stock Exchange
16:Method of DNA sequencing
141:whole genome sequencing
603:Life sciences industry
598:DNA sequencing methods
277:O2 is oxygen molecule;
156:
143:at affordable prices.
51:solid phase sequencing
154:
268:PPi is pyrophosphate
77:) and a nucleotide (
548:Metzker M. (2005).
567:10.1101/gr.3770505
287:is carbon dioxide;
157:
59:firefly luciferase
608:Molecular biology
502:"Biotage History"
430:(5375): 363–365.
353:genome sequencing
345:454 Life Sciences
327:was founded with
321:Pyrosequencing AB
315:Commercialization
301:chain termination
122:454 Life Sciences
118:Jonathan Rothberg
49:by combining the
615:
579:
569:
535:
534:
522:
516:
515:
513:
512:
497:
491:
490:
488:
486:
475:
469:
462:
456:
455:
415:
409:
403:
397:
391:
385:
379:
165:chemoluminescent
623:
622:
618:
617:
616:
614:
613:
612:
583:
582:
560:(12): 1767–76.
554:Genome Research
547:
544:
542:Further reading
539:
538:
524:
523:
519:
510:
508:
506:www.biotage.com
499:
498:
494:
484:
482:
477:
476:
472:
463:
459:
417:
416:
412:
404:
400:
392:
388:
380:
376:
371:
329:venture capital
325:Uppsala, Sweden
317:
305:genome assembly
297:
286:
259:
196:ATP sulfurylase
149:
126:microfabricated
86:Mostafa Ronaghi
71:ATP sulfurylase
22:is a method of
17:
12:
11:
5:
621:
619:
611:
610:
605:
600:
595:
585:
584:
581:
580:
543:
540:
537:
536:
531:Fierce Biotech
517:
492:
470:
457:
410:
398:
386:
373:
372:
370:
367:
316:
313:
309:repetitive DNA
296:
293:
292:
291:
288:
284:
281:
278:
275:
272:
269:
262:
261:
257:
254:
247:
246:
239:
231:
192:DNA polymerase
161:DNA polymerase
148:
145:
137:DNA sequencing
102:DNA polymerase
67:DNA polymerase
32:DNA polymerase
24:DNA sequencing
20:Pyrosequencing
15:
13:
10:
9:
6:
4:
3:
2:
620:
609:
606:
604:
601:
599:
596:
594:
593:Biotechnology
591:
590:
588:
577:
573:
568:
563:
559:
555:
551:
546:
545:
541:
532:
528:
521:
518:
507:
503:
496:
493:
481:
474:
471:
467:
461:
458:
453:
449:
445:
441:
437:
433:
429:
425:
421:
414:
411:
408:
402:
399:
396:
390:
387:
384:
378:
375:
368:
366:
364:
360:
358:
354:
350:
346:
342:
338:
334:
330:
326:
322:
314:
312:
310:
306:
302:
294:
289:
282:
279:
276:
273:
270:
267:
266:
265:
255:
252:
251:
250:
244:
240:
236:
232:
229:
228:pyrophosphate
225:
221:
217:
216:
215:
213:
209:
205:
201:
197:
193:
189:
185:
180:
177:
173:
169:
166:
162:
153:
146:
144:
142:
138:
134:
130:
127:
123:
119:
114:
111:
107:
103:
99:
95:
91:
90:Mathias Uhlen
87:
82:
80:
76:
72:
68:
64:
60:
56:
53:method using
52:
48:
44:
43:Mathias Uhlen
39:
37:
36:pyrophosphate
33:
29:
25:
21:
557:
553:
530:
520:
509:. Retrieved
505:
495:
483:. Retrieved
473:
460:
427:
423:
413:
401:
389:
377:
361:
357:metagenomics
351:, including
331:provided by
320:
319:The company
318:
298:
290:hv is light.
263:
248:
181:
158:
115:
83:
73:and firefly
55:streptavidin
40:
19:
18:
295:Limitations
176:nucleotides
28:nucleotides
587:Categories
511:2022-09-19
369:References
210:(APS) and
200:luciferase
129:microarray
106:luciferase
75:luciferase
500:Biotage.
333:HealthCap
212:luciferin
147:Procedure
139:allowing
94:Pål Nyren
47:Pål Nyren
576:16339375
485:4 August
478:QIAGEN.
452:26331871
133:genomics
108:and the
444:9705713
424:Science
264:where:
243:apyrase
204:apyrase
110:apyrase
98:apyrase
63:enzymes
574:
450:
442:
341:Qiagen
230:(PPi).
188:primer
168:enzyme
104:, the
448:S2CID
363:Roche
224:dNTPs
184:ssDNA
572:PMID
487:2017
440:PMID
355:and
202:and
92:and
79:dNTP
45:and
562:doi
432:doi
428:281
323:in
235:ATP
172:DNA
589::
570:.
558:15
556:.
552:.
529:.
504:.
446:.
438:.
426:.
422:.
359:.
283:CO
214:.
198:,
194:,
88:,
69:,
578:.
564::
533:.
514:.
489:.
468:;
454:.
434::
285:2
258:2
222:(
65:(
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.