1323:
203:
47:
2470:"we date the Y-chromosomal most recent common ancestor (MRCA) in Africa at 254 (95% CI 192–307) kya and detect a cluster of major non-African founder haplogroups in a narrow time interval at 47–52 kya, consistent with a rapid initial colonization model of Eurasia and Oceania after the out-of-Africa bottleneck. In contrast to demographic reconstructions based on mtDNA, we infer a second strong bottleneck in Y-chromosome lineages dating to the last 10 ky. We hypothesize that this bottleneck is caused by cultural changes affecting variance of reproductive success among males."
1194:. it has also been observed in Eastern, Central and Southern of Arabia. Current results, according to FTDNA, suggest that some branches such as A-V1127 originated in Arabia. Additionally, as suggested by experts as seen in TMRCA in Yfull tree, this haplogroup must have undergone a bottleneck time when people who represent this haplogroup suffered some sort of extinction and sharply decreased in number. Noteworthy, non semitic speakers don't have this haplogroup neither the koi-san or the nilots or the Cushites.
195:
5346:
975:
ancestral state of all known Y chromosome SNPs. To further characterize this lineage, which we dubbed A00, for proposed nomenclature)"; "We have renamed the basal branch in
Cruciani et al. as A0 (previously A1b) and refer to the presently reported lineage as A00. For deep branches discovered in the future, we suggest continuing the nomenclature A000, and so on." It has an estimated age of around 275 kya, so is roughly contemporary with the known appearance of earliest known
347:. According to Wood et al. (2005) and Rosa et al. (2007), such relatively recent population movements from West Africa changed the pre-existing population Y chromosomal diversity in Central, Southern and Southeastern Africa, replacing the previous haplogroups in these areas with the now dominant E1b1a lineages. Traces of ancestral inhabitants, however, can be observed today in these regions via the presence of the Y DNA haplogroups A-M91 and
2721:(2012) agreed with a plausible placement in "the north-western quadrant of the African continent" for the emergence of the A1b haplogroup: "the hypothesis of an origin in the north-western quadrant of the African continent for the A1b haplogroup, and, together with recent findings of ancient Y-lineages in central-western Africa, provide new evidence regarding the geographical origin of human MSY diversity".
1350:
new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
2675:, suggesting plausibility for the Y-MRCA living in the general region of North-Central Africa". In a sample of 2204 African Y-chromosomes, 8 chromosomes belonged to either haplogroup A1b or A1a. Haplogroup A1a was identified in two Moroccan Berbers, one Fulbe and one Tuareg people from Niger. Haplogroup A1b was identified in three Bakola pygmies from Southern Cameroon and one Algerian Berber.
1354:
4410:
others carried DYS19-15). These 11 chromosomes were all found in a sample of 174 (~6.3%) Mbo individuals from western
Cameroon (Figure 2). Seven of these Mbo chromosomes were available for further testing, and the genotypes were found to be identical at 37 of 39 SNPs known to be derived on the A00 chromosome (i.e., two of these genotyped SNPs were ancestral in the Mbo samples)".
2530:
Crevecoeur, Isabelle; de Maret, Pierre; Fomine, Forka Leypey Mathew; Lavachery, Philippe; Mindzie, Christophe Mbida; Orban, Rosine; Sawchuk, Elizabeth; Semal, Patrick; Thomas, Mark G.; Van Neer, Wim; Veeramah, Krishna R.; Kennett, Douglas J.; Patterson, Nick; Hellenthal, Garrett; Lalueza-Fox, Carles; MacEachern, Scott; Prendergast, Mary E.; Reich, David (30 January 2020).
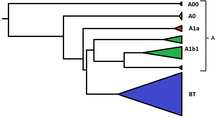
1376:). While the SNP marker M91 had been regarded as a key to identifying haplogroup BT, it was realised that the region surrounding M91 was a mutational hotspot, which is prone to recurrent back-mutations. Moreover, the 8T stretch of Haplogroup A represented the ancestral state of M91, and the 9T of haplogroup BT a derived state, which arose following the
3049:
Francalacci P, Morelli L, Angius A, Berutti R, Reinier F, Atzeni R, Pilu R, Busonero F, Maschio A, Zara I, Sanna D, Useli A, Urru MF, Marcelli M, Cusano R, Oppo M, Zoledziewska M, Pitzalis M, Deidda F, Porcu E, Poddie F, Kang HM, Lyons R, Tarrier B, Gresham JB, Li B, Tofanelli S, Alonso S, Dei M, Lai
4423:
Quotes: We can now clearly see that with 40% A00, the Bangwa represent the epicentre of A00 in this region, and very possibly in the world. As I shared in the last Lab Note, we found that so far there are two main subgroups of A00, defined by different Y-SNP mutations, which, naturally, divide along
1349:
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome
Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single
1238:
The subclade A1b1b2b (M13; formerly A3b2) is primarily distributed among
Nilotic populations in East Africa and northern Cameroon. It is different from the A subclades that are found in the Khoisan samples and only remotely related to them (it is actually only one of many subclades within haplogroup
245:
in 2013), have been detected in West Africa, Northwest Africa and
Central Africa. Cruciani et al. (2011) suggest that these lineages may have emerged somewhere in between Central and Northwest Africa. Scozzari et al. (2012) also supported "the hypothesis of an origin in the north-western quadrant of
2529:
Lipson, Mark; Ribot, Isabelle; Mallick, Swapan; Rohland, Nadin; Olalde, Iñigo; Adamski, Nicole; Broomandkhoshbacht, Nasreen; Lawson, Ann Marie; López, Saioa; Oppenheimer, Jonas; Stewardson, Kristin; Asombang, Raymond Neba’ane; Bocherens, Hervé; Bradman, Neil; Culleton, Brendan J.; Cornelissen, Els;
1380:
of 1T. This explained why subclades A1b and A1a, the deepest branches of
Haplogroup A, both possessed the 8T stretch. Similarly, the P97 marker, which was also used to identify haplogroup A, possessed the ancestral state in haplogroup A, but a derived state in haplogroup BT. Ultimately the tendency
4409:
Mendez et al. (2013), p. 455. Quote: "Upon searching a large pan-African database consisting of 5,648 samples from ten countries we identified 11 Y chromosomes that were invariant and identical to the A00 chromosome at five of the six Y-STRs (2 of the 11 chromosomes carried DYS19-16, whereas the
974:
Mendez et al. (2013) announced the discovery of a previously unknown haplogroup, for which they proposed the designator "A00". "Genotyping of a DNA sample that was submitted to a commercial genetic-testing facility demonstrated that the Y chromosome of this
African American individual carried the
6005:
K-M2313*, which as yet has no phylogenetic name, has been documented in two living individuals, who have ethnic ties to India and South East Asia. In addition, K-Y28299, which appears to be a primary branch of K-M2313, has been found in three living individuals from India. See: Poznik
315:
of modern humans) is believed to have originated. Estimates of its time depth have varied greatly, at either close to 190 kya or close to 140 kya in separate 2013 studies, and with the inclusion of the previously unknown "A00" haplogroup to about 270 kya in 2015 studies.
1141:
The subclade A1b1a1a (M6; formerly A2 and A1b1a1a-M6) is typically found among
Khoisan peoples. The authors of one study have reported finding haplogroup A-M6(xA-P28) in 28% (8/29) of a sample of Tsumkwe San and 16% (5/32) of a sample of
1129:
sharing the unusual surname Revis were identified as being from the A1a (M31) subclade. It was discovered that these men had a common male-line ancestor from the 18th century, but no previous information about
African ancestry was known.
4511:(2008). "Southern Sudanese" includes 26 Dinka, 15 Shilluk, and 12 Nuer. "Western Sudanese" includes 26 Borgu, 32 Masalit, and 32 Fur. "Northern Sudanese" includes 39 Nubians, 42 Beja, 33 Copts, 50 Gaalien, 28 Meseria, and 24 Arakien.
4323:
Mendez, Fernando L.; Krahn, Thomas; Schrack, Bonnie; Krahn, Astrid-Maria; Veeramah, Krishna R.; Woerner, August E.; Fomine, Forka Leypey Mathew; Bradman, Neil; Thomas, Mark G.; Karafet, Tatiana M.; Hammer, Michael F. (March 2013).
4273:
Mendez, Fernando L.; Krahn, Thomas; Schrack, Bonnie; Krahn, Astrid-Maria; Veeramah, Krishna R.; Woerner, August E.; Fomine, Forka Leypey Mathew; Bradman, Neil; Thomas, Mark G.; Karafet, Tatiana M.; Hammer, Michael F. (March 2013).
237:
lineages are also largely restricted to the San. However, the A lineages of
Southern Africa are sub-clades of A lineages found in other parts of Africa, suggesting that A sub-haplogroups arrived in Southern Africa from elsewhere.
1277:. Another study has reported finding haplogroup A3b2b-M118 in 6.8% (6/88) and haplogroup A3b2*-M13(xA3b2a-M171, A3b2b-M118) in 5.7% (5/88) of a mixed sample of Ethiopians, amounting to a total of 12.5% (11/88) A3b2-M13.
3766:
Shen P, Lavi T, Kivisild T, et al. (September 2004). "Reconstruction of patrilineages and matrilineages of Samaritans and other Israeli populations from Y-chromosome and mitochondrial DNA sequence variation".
5841:
1158:. The authors of another study have reported finding haplogroup A2 in 15.4% (6/39) of a sample of Khoisan males, including 5/39 A2-M6/M14/M23/M29/M49/M71/M135/M141(xA2a-M114) and 1/39 A2a-M114.
1357:
The revised y-chromosome family tree by Cruciani et al. 2011 compared with the family tree from Karafet et al. 2008. (The "A1a-T" shown here is now known as A1 and "A2-T" is now known as A1b.)
937:
A3a2 (A-M13; formerly A3b2), has been observed at very low frequencies in some Mediterranean islands. Without testing for any subclade, haplogroup A has been found in a sample of Greeks from
4132:
Di Giacomo F, Luca F, Anagnou N, et al. (September 2003). "Clinal patterns of human Y chromosomal diversity in continental Italy and Greece are dominated by drift and founder effects".
1381:
of M91 to back-mutate and (hence) its unreliability, led to M91 being discarded as a defining SNP by ISOGG in 2016. Conversely, P97 has been retained as a defining marker of Haplogroup BT.
3606:
Hassan HY, Underhill PA, Cavalli-Sforza LL, Ibrahim ME (November 2008). "Y-chromosome variation among Sudanese: restricted gene flow, concordance with language, geography, and history".
1002:(Bantu) (out of a sample of 174 (6.32%). Subsequent research suggested that the overall rate of A00 was even higher among the Mbo, i.e. 9.3% (8 of 86) were later found to fall within
2332:
218:; in other words, it is defined by the absence of the defining mutation of that group (M91). By this definition, haplogroup A includes all mutations that took place between the
5152:
Semino, O.; Passarino, G; Oefner, PJ; Lin, AA; et al. (2000), "The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans: A Y Chromosome Perspective",
4446:
Gonçalves R, Rosa A, Freitas A, et al. (November 2003). "Y-chromosome lineages in Cabo Verde Islands witness the diverse geographic origin of its first male settlers".
229:
Bearers of haplogroup A (i.e. absence of the defining mutation of haplogroup BT) have been found in Southern Africa's hunter-gatherer inhabited areas, especially among the
4627:
Semino O, Passarino G, Oefner PJ, et al. (November 2000). "The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective".
987:
man who had submitted his DNA for commercial genealogical analysis. The subsequent discovery of other males belonging to A00 led to the reclassification of Perry's Y as
4564:
4167:
Gonçalves R, Freitas A, Branco M, et al. (July 2005). "Y-chromosome lineages from Portugal, Madeira and Açores record elements of Sephardim and Berber ancestry".
3212:"Regional Differences in the Accumulation of SNPs on the Male-Specific Portion of the Human Y Chromosome Replicate Autosomal Patterns: Implications for Genetic Dating"
2589:"Regional Differences in the Accumulation of SNPs on the Male-Specific Portion of the Human Y Chromosome Replicate Autosomal Patterns: Implications for Genetic Dating"
983:. A00 is also sometimes known as "Perry's Y-chromosome" (or simply "Perry's Y"). This previously unknown haplogroup was discovered in 2012 in the Y chromosome of an
5868:
Van Oven M, Van Geystelen A, Kayser M, Decorte R, Larmuseau HD (2014). "Seeing the wood for the trees: a minimal reference phylogeny for the human Y chromosome".
5233:
Underhill, Peter A.; Shen, Peidong; Lin, Alice A.; Jin, Li; et al. (November 2000). "Y chromosome sequence variation and the history of human populations".
4056:"Supplementary Data: Signatures of the Preagricultural Peopling Processes in Sub-Saharan Africa as Revealed by the Phylogeography of Early Y Chromosome Lineages"
1344:
893:, occurring among 41% of one sample from this population (Cruciani et al. 2002). Elsewhere in the region, haplogroup A has been reported in 14.6% (7/48) of an
1024:
group of Cameroon (Grassfields Bantu): 27 of 67 (40.3%) samples were positive for A00a (L1149). One Bangwa individual did not fit into either A00a or A00b.
889:
Haplogroup A is found at low to moderate frequencies in the Horn of Africa. The clade is observed at highest frequencies among the 41% of a sample of the
1361:
Initial sequencing of the human Y-chromosome had suggested that first split in the Y-Chromosome family tree occurred with the mutations that separated
929:
In Asia, haplogroup A has been observed at low frequencies in Asia Minor and the Middle East among Aegean Turks, Palestinians, Jordanians, Yemenites.
710:
In a composite sample of 3551 African men, Haplogroup A had a frequency of 5.4%. The highest frequencies of haplogroup A have been reported among the
5287:
3815:"Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages"
149:
2418:(see there); including the A00 lineage, Karmin et al. (2015) and Trombetta et al. (2015) estimate ages of 254,000 and 291,000 ybp, respectively.
5846:
2587:
Trombetta, Beniamino; d'Atanasio, Eugenia; Massaia, Andrea; Myres, Natalie M.; Scozzari, Rosaria; Cruciani, Fulvio; Novelletto, Andrea (2015).
1322:
210:
Though there are terminological challenges to define it as a haplogroup, haplogroup A has come to mean "the foundational haplogroup" (viz. of
2833:"Signatures of the preagricultural peopling processes in sub-Saharan Africa as revealed by the phylogeography of early Y chromosome lineages"
4424:
ethnic lines: A00a among the Bangwa, and A00b among the Mbo. We also found the one Bangwa sample which didn't belong to either subgroup.".
4220:
Capelli C, Redhead N, Romano V, et al. (March 2006). "Population structure in the Mediterranean basin: a Y chromosome perspective".
3099:
Poznik GD, Henn BM, Yee MC, Sliwerska E, Euskirchen GM, Lin AA, Snyder M, Quintana-Murci L, Kidd JM, Underhill PA, Bustamante CD (2013).
226:) and the mutation defining haplogroup BT (estimated at some 140–150 kya), including any extant subclades that may yet to be discovered.
4372:
At first, via Mendez et al. (2013), this was announced as "extremely ancient" (95% confidence interval 237–581 kya for the age of the
5330:
2337:
2327:
141:
3660:
Underhill PA, Shen P, Lin AA, et al. (November 2000). "Y chromosome sequence variation and the history of human populations".
180:; these branches are only very distantly related, and are not more closely related to each other than they are to haplogroup BT.
5074:"Patterns of inter- and intra-group genetic diversity in the Vlax Roma as revealed by Y chromosome and mitochondrial DNA lineages"
6065:
4712:
1182:
The subclade (appropriately considered as a distinct haplogroup) A1b1b1 (M28; formerly A3a) has only been rarely observed in the
845:
815:
5320:
4925:
4419:
Which of Cameroon's peoples have members of haplogroup A00? // experiment.com update of funded research (Schrack/Fomine Forka)
953:, though they have not definitively excluded the possibility that either of these individuals may belong to a rare subclade of
2677:
Cruciani, Fulvio; Trombetta, Beniamino; Massaia, Andrea; Destro-Bisol, Giovanni; Sellitto, Daniele; Scozzari, Rosaria (2011).
202:
144:, which includes all living human Y chromosomes. Bearers of extant sub-clades of haplogroup A are almost exclusively found in
46:
5292:
3716:"A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes"
1086:
have reported finding A1a-M31 in 5.1% (14/276) of a sample from Guinea-Bissau and in 0.5% (1/201) of a pair of samples from
4739:
Scorrano, Gabriele; Viva, Serena; Pinotti, Thomaz; Fabbri, Pier Francesco; Rickards, Olga; Macciardi, Fabio (26 May 2022).
1369:
and haplogroup A more broadly. Subsequently, many intervening splits between Y-chromosomal Adam and BT, also became known.
1074:
The subclade A1a (M31) has been found in approximately 2.8% (8/282) of a pool of seven samples of various ethnic groups in
762:
Further downstream around the Nile valley, the subclade A3b2 has also been observed at very low frequencies in a sample of
4966:"A Predominantly Indigenous Paternal Heritage for the Austronesian-Speaking Peoples of Insular Southeast Asia and Oceania"
4935:
743:
In North Africa, haplogroup A is largely absent. Its subclade A1 has been observed at trace frequencies among Moroccans.
4390:
755:(53%), especially the Dinka Sudanese (61.5%). Haplogroup A3b2-M13 also has been observed in another sample of a South
4521:
Cinnioğlu C, King R, Kivisild T; et al. (January 2004). "Excavating Y-chromosome haplotype strata in Anatolia".
913:-speaking tribes with frequency ranging from 10% to 70%. This particular haplogroup was not found in a sample of the
3500:"Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes"
976:
5987:
Haplogroup K2b1 (P397/P399) is also known as Haplogroup MS, but has a broader and more complex internal structure.
4741:"Bioarchaeological and palaeogenomic portrait of two Pompeians that died during the eruption of Vesuvius in 79 AD"
4398:
328:
4884:"An African American paternal lineage adds an extremely ancient root to the human Y chromosome phylogenetic tree"
4326:"An African American Paternal Lineage Adds an Extremely Ancient Root to the Human Y Chromosome Phylogenetic Tree"
4276:"An African American Paternal Lineage Adds an Extremely Ancient Root to the Human Y Chromosome Phylogenetic Tree"
4714:
Genetic Patterns of Y-chromosome and Mitochondrial DNA Variation, with Implications to the Peopling of the Sudan
5509:
2365:
2355:
2350:
1995:
1934:
1568:
1319:
Haplogroup A-M13 has been found among three Neolithic period fossils excavated from the Kadruka site in Sudan.
949:. The authors of one study have reported finding what appears to be haplogroup A in 3.1% (2/65) of a sample of
332:
2395:
2385:
2380:
2375:
2370:
2360:
1873:
1812:
1751:
1690:
1629:
1446:
1166:
The clade A1b1b (M32; formerly A3) contains the most populous branches of haplogroup A and is mainly found in
5449:
2891:"A revised root for the human Y chromosomal phylogenetic tree: the origin of patrilineal diversity in Africa"
2679:"A Revised Root for the Human Y Chromosomal Phylogenetic Tree: The Origin of Patrilineal Diversity in Africa"
2390:
1507:
1210:/Sekele, 6/29 = 21% Tsumkwe San, 1/18 = 6% Dama). However, it also has been found with lower frequency among
6033:
Haplogroup S, as of 2017, is also known as K2b1a. (Previously the name Haplogroup S was assigned to K2b1a4.)
5712:
5439:
5190:"Y-Chromosome Evidence for a Northward Migration of Modern Humans into Eastern Asia during the Last Ice Age"
4055:
2134:
1187:
840:
Without testing for any subclade, haplogroup A Y-DNA has been observed in samples of several populations of
157:
6042:
Haplogroup M, as of 2017, is also known as K2b1b. (Previously the name Haplogroup M was assigned to K2b1d.)
5536:
5519:
4436:
et al. Ancient Human DNA from Shum Laka (Cameroon) in the Context of African Population History // SAA 2019
1009:
Further research in 2015 indicates that the modern population with the highest concentration of A00 is the
5701:
5693:
5433:
5298:
4964:
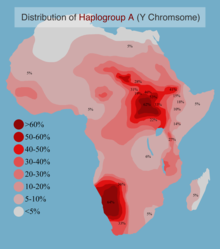
Capelli, Cristian; Wilson, James F.; Richards, Martin; Stumpf, Michael P.H.; et al. (February 2001).
999:
194:
5527:
3050:
S, Mulas A, Whalen MB, Uzzau S, Jones C, Schlessinger D, Abecasis GR, Sanna S, Sidore C, Cucca F (2013).
5763:
5609:
4558:
2309:
1362:
995:
5686:
5679:
5672:
5072:
Kaladjieva, Luba; Calafell, Francesc; Jobling, Mark A; Angelicheva, Dora; et al. (February 2001).
5282:
4672:"Isolates in a corridor of migrations: a high-resolution analysis of Y-chromosome variation in Jordan"
2723:
Scozzari R; Massaia A; D'Atanasio E; Myres NM; Perego UA; et al. (2012). Caramelli, David (ed.).
2057:
The following research teams per their publications were represented in the creation of the YCC Tree.
1280:
Haplogroup A-M13 also has been observed occasionally outside of Central and Eastern Africa, as in the
241:
The two most basal lineages of haplogroup A, A0 and A1 (prior to the announcement of the discovery of
5802:
5721:
5656:
5600:
5585:
5571:
5492:
5464:
5161:
5009:
Hammer, Michael F.; Karafet, Tatiana M.; Redd, Alan J.; Jarjanazi, Hamdi; et al. (1 July 2001).
4861:
4798:"New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree"
4752:
4636:
3401:
3223:
3112:
3063:
3013:
2954:
2736:
2600:
2543:
1377:
348:
336:
124:
31:
5795:
5790:
3346:"Genetic and demographic implications of the Bantu expansion: insights from human paternal lineages"
1090:. The authors of another study have reported finding haplogroup A1a-M31 in 5% (2/39) of a sample of
120:
116:
5313:
3002:"Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago"
1202:
The subclade A1b1b2a (M51; formerly A3b1) occurs most frequently among Khoisan peoples (6/11 = 55%
1146:/Sekele, and haplogroup A2b-P28 in 17% (5/29) of a sample of Tsumkwe San, 9% (3/32) of a sample of
214:); it is not defined by any mutation, but refers to any haplogroup which is not descended from the
3925:
Semino O, Santachiara-Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA (January 2002).
3541:"The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations"
3101:"Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females"
857:
849:
343:
from where it spread around 5,000 years ago to Central, Southern and Southeastern Africa with the
5893:
5731:
5405:
5388:
5258:
4546:
4481:
4255:
4202:
4111:
3979:"Africans in Yorkshire? The deepest-rooting clade of the Y phylogeny within an English genealogy"
3792:
3685:
3570:
1366:
1297:
161:
103:
87:
3871:"The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations"
30:
This article is about the human Y-DNA haplogroup. For the unrelated human mtDNA haplogroup, see
807:
5962:
5943:
F-Y27277, sometimes known as F2'4, is both the parent clade of F2 and F4 and a child of F-M89.
5885:
5823:
5818:
5809:
5752:
5743:
5663:
5634:
5594:
5552:
5504:
5422:
5250:
5221:
5177:
5140:
5109:"Paternal Population History of East Asia: Sources, Patterns, and Microevolutionary Processes"
5095:
5061:
5032:
4997:
4913:
4827:
4778:
4693:
4652:
4609:
4538:
4473:
4386:
4355:
4305:
4247:
4194:
4149:
4103:
4095:
4087:
4008:
3956:
3900:
3844:
3784:
3745:
3677:
3623:
3562:
3521:
3429:
3367:
3305:
3251:
3190:
3138:
3089:
3031:
2982:
2920:
2864:
2813:
2764:
2708:
2628:
2569:
2511:
2461:
1079:
910:
1219:
5877:
5785:
5543:
5497:
5242:
5211:
5201:
5169:
5130:
5120:
5085:
5053:
5022:
4987:
4977:
4903:
4895:
4817:
4809:
4768:
4760:
4683:
4644:
4599:
4591:
4530:
4463:
4455:
4345:
4337:
4295:
4287:
4237:
4229:
4184:
4176:
4141:
4077:
4067:
3998:
3990:
3946:
3938:
3890:
3882:
3834:
3826:
3776:
3735:
3727:
3669:
3615:
3552:
3511:
3419:
3409:
3357:
3295:
3285:
3241:
3231:
3180:
3172:
3128:
3120:
3079:
3071:
3021:
2972:
2962:
2910:
2902:
2854:
2844:
2803:
2795:
2754:
2744:
2698:
2690:
2668:
2618:
2608:
2559:
2551:
2501:
2493:
2451:
2443:
1010:
984:
73:
339:
in general is believed to have originated in Northeast Africa, and was later introduced to
5557:
5027:
5010:
4929:
2434:
1330:
Haplogroup A-M13 was also found in a male victim of the Mt. Vesuvius eruption in Pompeii.
1215:
1171:
1091:
344:
292:
288:
4844:
3161:"A recent bottleneck of Y chromosome diversity coincides with a global change in culture"
3052:"Low-pass DNA sequencing of 1200 Sardinians reconstructs European Y-chromosome phylogeny"
2784:"A recent bottleneck of Y chromosome diversity coincides with a global change in culture"
2430:"A recent bottleneck of Y chromosome diversity coincides with a global change in culture"
1015:
5359:
Please help update this article to reflect recent events or newly available information.
5165:
4756:
4640:
3405:
3227:
3116:
3067:
3017:
2958:
2889:
Cruciani F, Trombetta B, Massaia A, Destro-Bisol G, Sellitto D, Scozzari R (June 2011).
2740:
2604:
2547:
17:
5773:
5566:
5399:
5306:
5216:
5189:
5135:
5108:
4992:
4965:
4908:
4883:
4822:
4797:
4773:
4740:
4604:
4579:
4394:
4350:
4325:
4300:
4275:
4003:
3978:
3895:
3870:
3839:
3814:
3557:
3540:
3424:
3390:"Y-chromosomal diversity in the population of Guinea-Bissau: a multiethnic perspective"
3389:
3300:
3273:
3246:
3211:
3185:
3160:
3133:
3100:
3084:
3051:
2977:
2942:
2915:
2890:
2808:
2783:
2759:
2724:
2703:
2678:
2672:
2623:
2588:
2564:
2531:
2506:
2481:
2456:
2429:
1183:
1167:
1043:
The haplogroup names "A-V148" and "A-CTS2809/L991" refer to the exact same haplogroup.
1021:
559:
469:
431:
304:
300:
242:
99:
77:
5910:
5107:
Karafet, Tatiana; Xu, Liping; Du, Ruofu; Wang, William; et al. (September 2001).
5057:
4718:
4145:
3951:
3926:
3740:
3715:
3327:
2943:"Molecular dissection of the basal clades in the human Y chromosome phylogenetic tree"
2725:"Molecular Dissection of the Basal Clades in the Human Y Chromosome Phylogenetic Tree"
6059:
5966:
5648:
5642:
5628:
5617:
5580:
5483:
5478:
5469:
5455:
5416:
5188:
Su, Bing; Xiao, Junhua; Underhill, Peter; Deka, Ranjan; et al. (December 1999).
4233:
4180:
3927:"Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny"
1289:
1281:
1270:
1211:
1155:
1107:
1075:
958:
954:
894:
803:
802:
Haplogroup A3b2-M13 has been observed in populations of northern Cameroon (2/9 = 22%
775:
719:
685:
672:
646:
620:
584:
533:
266:
250:
234:
215:
211:
153:
128:
112:
108:
5897:
5277:
5262:
4922:
4578:
Nebel A, Filon D, Brinkmann B, Majumder PP, Faerman M, Oppenheim A (November 2001).
4550:
4485:
4259:
4115:
3689:
2163:(A4987/YP3666, A4981, A4982/YP2683, A4984/YP2995, A4985/YP3292, A4986, A4988/YP3731)
4580:"The Y chromosome pool of Jews as part of the genetic landscape of the Middle East"
4206:
3796:
3574:
3272:
Abu-Amero KK, Hellani A, González AM, Larruga JM, Cabrera VM, Underhill PA (2009).
2415:
1372:
A major shift in the understanding of the Y-DNA tree came with the publication of (
1274:
1259:
1223:
1207:
1147:
1143:
1099:
1047:
980:
898:
878:
861:
659:
607:
481:
278:
254:
5173:
4648:
1035:
pygmies. One individual carried the deeply divergent Y chromosome haplogroup A00.
1027:
Geneticists sequenced genome-wide DNA data from four people buried at the site of
319:
The clade has also been observed at notable frequencies in certain populations in
249:
Haplogroup A1b1b2 has been found among ancient fossils excavated at Balito Bay in
4670:
Flores C, Maca-Meyer N, Larruga JM, Cabrera VM, Karadsheh N, Gonzalez AM (2005).
4020:
3236:
2967:
2749:
2613:
1269:, one study has reported finding haplogroup A-M13 in 14.6% (7/48) of a sample of
4796:
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF (2008).
2664:
1353:
1247:
1227:
1203:
1151:
917:
from Tanzania, a population sometimes proposed as a remnant of a Late Stone Age
890:
881:
in Tanzania (3/43 = 7.0% (Luis et al. 2004) to 1/6 = 17% (Knight et al. 2003)).
853:
823:
819:
752:
715:
633:
546:
507:
494:
405:
340:
4939:
4899:
4764:
4341:
4291:
3274:"Saudi Arabian Y-Chromosome diversity and its relationship with nearby regions"
2906:
2694:
2497:
4688:
4671:
4534:
4459:
2555:
1095:
1087:
1031:
in Cameroon between 8000–3000 years ago, who were most genetically similar to
787:
520:
230:
223:
6018:
4099:
4091:
3321:
3319:
833:
Haplogroup A-M91(xA1a-M31, A2-M6/M14/P3/P4, A3-M32) has been observed in the
5090:
5073:
5044:
Jobling, Mark A.; Tyler-Smith, Chris (2000), "New uses for new haplotypes",
4072:
3994:
3516:
3499:
3414:
3362:
3345:
3290:
3124:
3075:
3026:
3001:
2849:
2832:
2532:"Ancient West African foragers in the context of African population history"
1122:
1028:
918:
763:
730:
1 African American Male out of Lacrosse, WI USA, Moses, Ramon, A00, A00-AF8
382:
198:
Tree showing relationship between branches of haplogroup A and haplogroup BT
5889:
5254:
5225:
5181:
5144:
5099:
5065:
5036:
5001:
4917:
4831:
4782:
4697:
4656:
4613:
4542:
4477:
4359:
4309:
4251:
4198:
4153:
4107:
4012:
3960:
3904:
3848:
3788:
3749:
3681:
3627:
3566:
3525:
3433:
3371:
3309:
3255:
3194:
3142:
3093:
3035:
2986:
2924:
2868:
2817:
2768:
2712:
2632:
2573:
2515:
2465:
945:
and in samples of Portuguese from southern Portugal, central Portugal, and
3830:
3176:
2799:
2447:
1190:
from Ethiopia, 1.1% (1/88) of a sample of Ethiopians, and 0.5% (1/201) in
4468:
4189:
4082:
4024:
2859:
1301:
1266:
1051:
938:
572:
320:
5925:
Haplogroup A0-T is also known as A-L1085 (and previously as A0'1'2'3'4).
4952:(chart highlighting new branches added to the A phylotree in March 2013)
5881:
4813:
3780:
3619:
1191:
1126:
1111:
1059:
1055:
946:
914:
711:
457:
356:
308:
296:
285:
6011:
5965:
was known as "Haplogroup K2". That name has since been re-assigned to
4242:
6022:, 2017, "Details of the Y-SNP markers included in the minimal Y tree"
4373:
1305:
1285:
1255:
1106:. Haplogroup A1a-M31 also has been found in 3% (2/64) of a sample of
950:
942:
834:
759:
population at a frequency of 45% (18/40), including 1/40 A3b2a-M171.
312:
303:. However, haplogroup A's oldest sub-clades are exclusively found in
274:
270:
219:
189:
145:
5978:
Haplogroup K2b (M1221/P331/PF5911) is also known as Haplogroup MPS.
5206:
5125:
4982:
4595:
3942:
3886:
3731:
1114:
and 2.3% (1/44) of a sample of unspecified ethnic affiliation from
206:
Projected spatial frequency distribution of haplogroup A in Africa.
5246:
4420:
3673:
2130:
1352:
1321:
1309:
1293:
1243:
1063:
1032:
841:
827:
811:
779:
756:
698:
418:
352:
324:
201:
193:
2831:
Batini C, Ferri G, Destro-Bisol G, et al. (September 2011).
1313:
1251:
1115:
1103:
1082:(5/64 = 7.8%). In an earlier study published in 2003, Gonçalves
783:
444:
5302:
3539:
Underhill PA, Passarino G, Lin AA, et al. (January 2001).
273:, though a very small handful of bearers have been reported in
5339:
5011:"Hierarchical Patterns of Global Human Y-Chromosome Diversity"
3813:
Cruciani F, Trombetta B, Sellitto D, et al. (July 2010).
3344:
Berniell-Lee G, Calafell F, Bosch E, et al. (July 2009).
2646:
2482:"The Divergence of Neandertal and Modern Human Y Chromosomes"
3714:
Cruciani F, Santolamazza P, Shen P, et al. (May 2002).
909:
One 2005 study has found haplogroup A in samples of various
257:, which have been dated to around 2149-1831 BP (2/2; 100%).
3977:
King TE, Parkin EJ, Swinfield G, et al. (March 2007).
901:
sample, and 13.6% (12/88) of another sample from Ethiopia.
351:
that are common in certain relict populations, such as the
5911:
International Society of Genetic Genealogy (ISOGG; 2015),
4865:
4389:
born in the United States between ca. 1819–1827, lived in
3869:
Luis JR, Rowold DJ, Regueiro M, et al. (March 2004).
3388:
Rosa A, Ornelas C, Jobling MA, Brehm A, Villems R (2007).
107:(Subclades of these include haplogroups A00a, A00b, A00c,
5952:
Haplogroup LT (L298/P326) is also known as Haplogroup K1.
4882:
Mendez FL, Krahn T, Schrack B, et al. (March 2013).
2941:
Scozzari R, Massaia A, D'Atanasio E, et al. (2012).
1326:
Geographical frequency distribution of Haplogroup A3-M13.
284:
The clade achieves its highest modern frequencies in the
3000:
Carina M. Schlebusch; et al. (28 September 2017).
2782:
Kamin M, Saag L, Vincente M, et al. (April 2015).
2065:
57:
270,000 BP, 275,000 BP (303,000-241,000 BP), 291,000 BP
3498:
Wood ET, Stover DA, Ehret C, et al. (July 2005).
2333:
Y-DNA haplogroups in populations of Sub-Saharan Africa
1246:, haplogroup A-M13 has been found in 28/53 = 52.8% of
1133:
In 2023, Lacrosse, WI, 1 Male, A1a-M31, Moses, Ramon.
4376:
including the lineage of this postulated haplogroup).
1222:, 3/53 = 6% non-Khoisan Southern Africans, 4/80 = 5%
2663:
2011, the most basal lineages have been detected in
5381:
3339:
3337:
3155:
3153:
94:
83:
69:
61:
53:
39:
1239:A). This finding suggests an ancient divergence.
4127:
4125:
3655:
3653:
3651:
3649:
3647:
3645:
3643:
3641:
3639:
3637:
3267:
3265:
1186:. In 5% (1/20) of a mixed sample of speakers of
994:Researchers later found A00 was possessed by 11
246:the African continent for the A1b haplogroup".
233:. In addition, the most basal mitochondrial DNA
4711:Yousif, Hisham; Eltayeb, Muntaser (July 2009).
3808:
3806:
3709:
3707:
3705:
3703:
3701:
3699:
3206:
3204:
2936:
2934:
2884:
2882:
2880:
2878:
2157:(L1149, FGC25576, FGC26292, FGC26293, FGC27741)
6024:(Access date of these pages: 9 December 2017)
4054:Batini, Chiara; et al. (September 2011).
5314:
3920:
3918:
3916:
3914:
3600:
3598:
3596:
3594:
3592:
3590:
3588:
3586:
3584:
1345:Conversion table for Y chromosome haplogroups
8:
4563:: CS1 maint: multiple names: authors list (
3493:
3491:
3489:
3487:
3485:
3483:
3481:
3479:
3477:
3475:
3473:
3471:
3469:
3467:
3465:
3463:
3326:International Society of Genetic Genealogy.
2414:equivalent to an estimate of the age of the
2129:The above phylogenetic tree is based on the
877:Bantus in Kenya (14%, Luis et al. 2004) and
327:groups in Central Africa, and less commonly
5996:Haplogroup P (P295) is also klnown as K2b2.
3864:
3862:
3860:
3858:
3761:
3759:
3461:
3459:
3457:
3455:
3453:
3451:
3449:
3447:
3445:
3443:
3383:
3381:
1304:(1/77 = 1.3%, 1/22 = 4.5%), the capital of
5321:
5307:
5299:
2069:
45:
5215:
5205:
5134:
5124:
5089:
5026:
4991:
4981:
4907:
4821:
4772:
4687:
4603:
4467:
4349:
4299:
4241:
4188:
4081:
4071:
4002:
3950:
3894:
3838:
3739:
3556:
3515:
3423:
3413:
3361:
3299:
3289:
3245:
3235:
3184:
3147:Cruciani et al. (2011) estimated 142 kya.
3132:
3083:
3025:
2976:
2966:
2914:
2858:
2848:
2807:
2758:
2748:
2702:
2622:
2612:
2563:
2505:
2455:
2077:
1262:, and 5/216 = 2.3% of Northern Sudanese.
837:people of southern Cameroon (3/33 = 9%).
269:", it is almost completely restricted to
220:Y-chromosomal most recent common ancestor
164:. The known branches of haplogroup A are
5934:Haplogroup A1 is also known as A1'2'3'4.
5278:Family Tree DNA — Y-Haplogroup A Project
1383:
1373:
751:Haplogroup A3b2-M13 is common among the
5860:
3972:
3970:
2407:
2117:
2093:
156:, bearers of which participated in the
5194:The American Journal of Human Genetics
5113:The American Journal of Human Genetics
4970:The American Journal of Human Genetics
4556:
4330:The American Journal of Human Genetics
4280:The American Journal of Human Genetics
2683:The American Journal of Human Genetics
2486:The American Journal of Human Genetics
2101:
2085:
265:By definition of haplogroup A as "non-
65:275,000 BP (split with other lineages)
36:
5028:10.1093/oxfordjournals.molbev.a003906
2137:, and subsequent published research.
844:, including 9% (3/33) of a sample of
7:
5847:Y-DNA haplogroups of historic people
5822:
5817:
5808:
5806:
5801:
5799:
5794:
5789:
5784:
5778:
5771:
5761:
5759:
5749:
5741:
5729:
5727:
5718:
5710:
5668:
5661:
5655:
5653:
5647:
5641:
5639:
5633:
5627:
5615:
5605:
5599:
5593:
5584:
5579:
5570:
5565:
5556:
5551:
5515:
5501:
5384:
331:speakers, who largely belong to the
5489:
5429:
5412:
5283:African Haplogroup project at FTDNA
1254:of central Sudan, 25/90 = 27.8% of
5078:European Journal of Human Genetics
4936:"Y-Haplogroup A Phylogenetic Tree"
3604:28/53 (Dinka, Nuer, and Shilluk),
3558:10.1046/j.1469-1809.2001.6510043.x
2109:
1206:, 11/39 = 28% Khoisan, 7/32 = 22%
1150:/Sekele, 9% (1/11) of a sample of
25:
5916:. (Access date: 1 February 2015.)
4399:FamilyTreeDNA, Haplogroup A chart
4021:"Yorkshire clan linked to Africa"
2338:Y-DNA haplogroups by ethnic group
2328:Human Y-chromosome DNA haplogroup
311:, where it (and by extension the
142:human Y-chromosome DNA haplogroup
27:Human Y-chromosome DNA haplogroup
5344:
4234:10.1111/j.1529-8817.2005.00224.x
4181:10.1111/j.1529-8817.2005.00161.x
2480:Mendez, L.; et al. (2016).
1273:and 10.3% (8/78) of a sample of
791:
5969:, the sibling of Haplogroup LT.
5015:Molecular Biology and Evolution
4060:Molecular Biology and Evolution
1154:, and 6% (1/18) of a sample of
860:, and 2% (1/60) of a sample of
786:, and one male individual from
212:contemporary human populations
1:
5174:10.1126/science.290.5494.1155
5058:10.1016/S0168-9525(00)02057-6
4957:Sources for conversion tables
4649:10.1126/science.290.5494.1155
4146:10.1016/S1055-7903(03)00016-2
2217:(L602, V50, V82, V198, V224)
1296:(4/147 = 2.7%, 3/92 = 3.3%),
1098:and 2% (1/55) of a sample of
5331:Y-chromosome DNA haplogroups
4938:. March 2013. Archived from
3237:10.1371/journal.pone.0134646
2968:10.1371/journal.pone.0049170
2750:10.1371/journal.pone.0049170
2614:10.1371/journal.pone.0134646
2428:Karmin; et al. (2015).
2066:Jobling and Tyler-Smith 2000
778:, two male individuals from
774:Eight male individuals from
5329:Phylogenetic tree of human
4850:. (Access: 29 August 2017.)
897:sample, 10.3% (8/78) of an
856:, 2% (1/57) of a sample of
852:, 2% (1/46) of a sample of
848:, 3% (1/36) of a sample of
782:, one male individual from
295:, followed closely by many
6082:
5913:Y-DNA Haplogroup Tree 2015
5815:
5782:
5739:
5708:
5699:
5691:
5684:
5677:
5670:
5625:
5591:
5577:
5563:
5549:
5533:
5517:
5386:
4900:10.1016/j.ajhg.2013.02.002
4847:Y-DNA Haplogroup Tree 2016
4765:10.1038/s41598-022-10899-1
4342:10.1016/j.ajhg.2013.02.002
4292:10.1016/j.ajhg.2013.02.002
2907:10.1016/j.ajhg.2011.05.002
2695:10.1016/j.ajhg.2011.05.002
2498:10.1016/j.ajhg.2016.02.023
2240:formerly A1b1a1a1b and A2b
2189:formerly A1a-T, A0 and A1b
1387:YCC 2002/2008 (Shorthand)
1342:
977:anatomically modern humans
187:
29:
5836:
5614:
5607:
5541:
5524:
5508:
5503:
5496:
5491:
5482:
5477:
5475:
5468:
5463:
5461:
5454:
5447:
5445:
5438:
5431:
5421:
5414:
5404:
5397:
5395:
5379:
5353:This article needs to be
5337:
4689:10.1007/s10038-005-0274-4
4535:10.1007/s00439-003-1031-4
4460:10.1007/s00439-003-1007-4
4044:16/26, Hassan et al. 2008
2556:10.1038/s41586-020-1929-1
369:
44:
4717:(Thesis). Archived from
1258:, 4/32 = 12.5% of local
1121:In 2007, seven men from
941:on the Aegean island of
364:Haplogroup A frequencies
70:Possible place of origin
18:Macro-haplogroup A(X-BT)
6066:Human Y-DNA haplogroups
5961:Between 2002 and 2008,
5091:10.1038/sj.ejhg.5200597
4864:. FTDNA. Archived from
3995:10.1038/sj.ejhg.5201771
3517:10.1038/sj.ejhg.5201408
3415:10.1186/1471-2148-7-124
3328:"Y-DNA Haplogroup Tree"
3291:10.1186/1471-2156-10-59
3125:10.1126/science.1237619
3076:10.1126/science.1237947
3027:10.1126/science.aao6266
1250:, 13/28 = 46.4% of the
1188:South Semitic languages
1078:, especially among the
1062:at 1.5%. Also found in
222:(estimated at some 270
158:Out of Africa migration
54:Possible time of origin
5288:Spread of Haplogroup A
3608:Am. J. Phys. Anthropol
2659:According to Cruciani
1358:
1327:
1218:, including 2/28 = 7%
1080:Papel-Manjaco-Mancanha
395:Tsumkwe San (Namibia)
207:
199:
4134:Mol. Phylogenet. Evol
4073:10.1093/molbev/msr089
3831:10.1038/ejhg.2009.231
3363:10.1093/molbev/msp069
3222:(7): e0134646. 2015.
3177:10.1101/gr.186684.114
2850:10.1093/molbev/msr089
2800:10.1101/gr.186684.114
2448:10.1101/gr.186684.114
1420:YCC 2010r (Longhand)
1356:
1325:
205:
197:
1417:YCC 2008 (Longhand)
1414:YCC 2005 (Longhand)
1411:YCC 2002 (Longhand)
1339:Phylogenetic history
1046:A0 is found only in
714:of Southern Africa,
313:patrilinear ancestor
152:), in contrast with
32:Haplogroup A (mtDNA)
5842:Y-DNA by population
5757:
5293:National Geographic
5166:2000Sci...290.1155S
4757:2022NatSR..12.6468S
4641:2000Sci...290.1155S
3406:2007BMCEE...7..124R
3228:2015PLoSO..1034646T
3171:(4): 459–66. 2015.
3117:2013Sci...341..562P
3068:2013Sci...341..565F
3018:2017Sci...358..652S
2959:2012PLoSO...749170S
2741:2012PLoSO...749170S
2605:2015PLoSO..1034646T
2548:2020Natur.577..665L
873:African Great Lakes
597:Mandara (Cameroon)
366:
162:early modern humans
5882:10.1002/humu.22468
5608:
5389:Y-chromosomal Adam
5046:Trends in Genetics
4928:2019-09-24 at the
4814:10.1101/gr.7172008
4745:Scientific Reports
3983:Eur. J. Hum. Genet
3819:Eur. J. Hum. Genet
3781:10.1002/humu.20077
3620:10.1002/ajpa.20876
3504:Eur. J. Hum. Genet
2142:Y-chromosomal Adam
2125:Phylogenetic trees
1367:Y-chromosomal Adam
1359:
1328:
362:
323:, as well as some
208:
200:
6053:
6052:
6015:, 2017, "K-M2335"
6013:YFull YTree v5.08
5963:Haplogroup T-M184
5831:
5830:
5776:
5766:
5756:
5747:
5735:
5724:
5716:
5704:
5696:
5689:
5682:
5675:
5620:
5539:
5531:
5522:
5452:
5436:
5419:
5402:
5374:
5373:
4942:on 18 August 2018
4888:Am. J. Hum. Genet
4584:Am. J. Hum. Genet
4507:Hisham Y. Hassan
3931:Am. J. Hum. Genet
3875:Am. J. Hum. Genet
3720:Am. J. Hum. Genet
3111:(6145): 562–565.
3062:(6145): 565–569.
3012:(6363): 652–655.
2895:Am. J. Hum. Genet
2542:(7792): 665–670.
2344:Y-DNA A subclades
2055:
2054:
1298:Palestinian Arabs
1248:Southern Sudanese
753:Southern Sudanese
708:
707:
379:Study population
135:
134:
16:(Redirected from
6073:
6043:
6040:
6034:
6031:
6025:
6003:
5997:
5994:
5988:
5985:
5979:
5976:
5970:
5959:
5953:
5950:
5944:
5941:
5935:
5932:
5926:
5923:
5917:
5908:
5902:
5901:
5865:
5772:
5762:
5750:
5742:
5730:
5720:
5711:
5700:
5692:
5685:
5678:
5671:
5616:
5535:
5526:
5518:
5448:
5432:
5415:
5398:
5382:
5369:
5366:
5360:
5348:
5347:
5340:
5323:
5316:
5309:
5300:
5266:
5229:
5219:
5209:
5200:(6): 1718–1724.
5184:
5160:(5494): 1155–9,
5148:
5138:
5128:
5103:
5093:
5068:
5040:
5030:
5021:(7): 1189–1203.
5005:
4995:
4985:
4951:
4949:
4947:
4921:
4911:
4870:
4869:
4857:
4851:
4842:
4836:
4835:
4825:
4793:
4787:
4786:
4776:
4736:
4730:
4729:
4727:
4726:
4708:
4702:
4701:
4691:
4667:
4661:
4660:
4635:(5494): 1155–9.
4624:
4618:
4617:
4607:
4575:
4569:
4568:
4562:
4554:
4518:
4512:
4505:
4499:
4498:23andme raw data
4496:
4490:
4489:
4471:
4443:
4437:
4431:
4425:
4421:available online
4417:
4411:
4407:
4401:
4385:Albert Perry, a
4383:
4377:
4370:
4364:
4363:
4353:
4320:
4314:
4313:
4303:
4270:
4264:
4263:
4245:
4228:(Pt 2): 207–25.
4217:
4211:
4210:
4192:
4175:(Pt 4): 443–54.
4164:
4158:
4157:
4129:
4120:
4119:
4085:
4075:
4066:(9): 2603–2613.
4051:
4045:
4042:
4036:
4035:
4033:
4032:
4016:
4006:
3974:
3965:
3964:
3954:
3922:
3909:
3908:
3898:
3866:
3853:
3852:
3842:
3810:
3801:
3800:
3763:
3754:
3753:
3743:
3711:
3694:
3693:
3657:
3632:
3631:
3602:
3579:
3578:
3560:
3536:
3530:
3529:
3519:
3495:
3438:
3437:
3427:
3417:
3385:
3376:
3375:
3365:
3341:
3332:
3331:
3323:
3314:
3313:
3303:
3293:
3269:
3260:
3259:
3249:
3239:
3208:
3199:
3198:
3188:
3157:
3148:
3146:
3136:
3097:
3087:
3046:
3040:
3039:
3029:
2997:
2991:
2990:
2980:
2970:
2938:
2929:
2928:
2918:
2886:
2873:
2872:
2862:
2852:
2828:
2822:
2821:
2811:
2779:
2773:
2772:
2762:
2752:
2716:
2706:
2657:
2651:
2650:
2643:
2637:
2636:
2626:
2616:
2584:
2578:
2577:
2567:
2526:
2520:
2519:
2509:
2477:
2471:
2469:
2459:
2425:
2419:
2412:
1384:
1316:(1/121 = 0.8%).
1312:(1/101=1%), and
1300:(2/143 = 1.4%),
1256:Western Sudanese
1226:, and 1/29 = 3%
1019:
1000:Western Cameroon
985:African-American
388:
367:
361:
355:Pygmies and the
309:Northwest Africa
150:African diaspora
74:Northwest Africa
49:
37:
21:
6081:
6080:
6076:
6075:
6074:
6072:
6071:
6070:
6056:
6055:
6054:
6049:
6048:
6047:
6046:
6041:
6037:
6032:
6028:
6004:
6000:
5995:
5991:
5986:
5982:
5977:
5973:
5960:
5956:
5951:
5947:
5942:
5938:
5933:
5929:
5924:
5920:
5909:
5905:
5867:
5866:
5862:
5857:
5851:
5832:
5375:
5370:
5364:
5361:
5358:
5349:
5345:
5333:
5327:
5274:
5269:
5235:Nature Genetics
5232:
5187:
5151:
5106:
5071:
5043:
5008:
4963:
4959:
4945:
4943:
4934:
4930:Wayback Machine
4881:
4878:
4873:
4860:Krahn, Thomas.
4859:
4858:
4854:
4843:
4839:
4802:Genome Research
4795:
4794:
4790:
4738:
4737:
4733:
4724:
4722:
4710:
4709:
4705:
4669:
4668:
4664:
4626:
4625:
4621:
4590:(5): 1095–112.
4577:
4576:
4572:
4555:
4520:
4519:
4515:
4506:
4502:
4497:
4493:
4445:
4444:
4440:
4432:
4428:
4418:
4414:
4408:
4404:
4384:
4380:
4371:
4367:
4322:
4321:
4317:
4272:
4271:
4267:
4222:Ann. Hum. Genet
4219:
4218:
4214:
4169:Ann. Hum. Genet
4166:
4165:
4161:
4131:
4130:
4123:
4053:
4052:
4048:
4043:
4039:
4030:
4028:
4019:
4017:
3976:
3975:
3968:
3924:
3923:
3912:
3868:
3867:
3856:
3812:
3811:
3804:
3765:
3764:
3757:
3726:(5): 1197–214.
3713:
3712:
3697:
3659:
3658:
3635:
3605:
3603:
3582:
3551:(Pt 1): 43–62.
3545:Ann. Hum. Genet
3538:
3537:
3533:
3497:
3496:
3441:
3387:
3386:
3379:
3350:Mol. Biol. Evol
3343:
3342:
3335:
3325:
3324:
3317:
3271:
3270:
3263:
3210:
3209:
3202:
3165:Genome Research
3159:
3158:
3151:
3098:
3048:
3047:
3043:
2999:
2998:
2994:
2940:
2939:
2932:
2888:
2887:
2876:
2837:Mol. Biol. Evol
2830:
2829:
2825:
2788:Genome Research
2781:
2780:
2776:
2722:
2676:
2658:
2654:
2645:
2644:
2640:
2599:(7): e0134646.
2586:
2585:
2581:
2528:
2527:
2523:
2479:
2478:
2474:
2435:Genome Research
2427:
2426:
2422:
2413:
2409:
2405:
2400:
2324:
2297:A1b1b2b1 (M118)
2283:A1b1b2a1 (P291)
2178:(CTS2809/L991)
2145:
2127:
2122:
2070:Kaladjieva 2001
1347:
1341:
1336:
1288:(2/30 = 6.7%),
1236:
1234:A1b1b2b (A-M13)
1216:Southern Africa
1200:
1198:A1b1b2a (A-M51)
1180:
1172:Southern Africa
1164:
1139:
1072:
1041:
1013:
972:
967:
935:
927:
907:
905:Southern Africa
887:
875:
870:
800:
772:
749:
741:
736:
728:
623:(South Africa)
386:
385:
345:Bantu expansion
293:Southern Africa
291:populations of
289:hunter-gatherer
263:
192:
186:
106:
62:Coalescence age
35:
28:
23:
22:
15:
12:
11:
5:
6079:
6077:
6069:
6068:
6058:
6057:
6051:
6050:
6045:
6044:
6035:
6026:
5998:
5989:
5980:
5971:
5954:
5945:
5936:
5927:
5918:
5903:
5870:Human Mutation
5859:
5858:
5853:
5852:
5850:
5849:
5844:
5838:
5837:
5834:
5833:
5829:
5828:
5826:
5821:
5816:
5813:
5812:
5807:
5805:
5800:
5798:
5793:
5788:
5783:
5780:
5779:
5777:
5770:
5768:
5760:
5758:
5748:
5740:
5737:
5736:
5728:
5726:
5717:
5709:
5706:
5705:
5698:
5690:
5683:
5676:
5669:
5667:
5660:
5654:
5652:
5646:
5640:
5638:
5632:
5626:
5623:
5622:
5613:
5606:
5604:
5598:
5592:
5589:
5588:
5583:
5578:
5575:
5574:
5569:
5564:
5561:
5560:
5555:
5550:
5547:
5546:
5540:
5532:
5523:
5516:
5513:
5512:
5507:
5502:
5500:
5495:
5490:
5487:
5486:
5481:
5476:
5473:
5472:
5467:
5462:
5459:
5458:
5453:
5446:
5443:
5442:
5437:
5430:
5427:
5426:
5420:
5413:
5410:
5409:
5403:
5396:
5393:
5392:
5385:
5380:
5377:
5376:
5372:
5371:
5352:
5350:
5343:
5338:
5335:
5334:
5328:
5326:
5325:
5318:
5311:
5303:
5297:
5296:
5285:
5280:
5273:
5272:External links
5270:
5268:
5267:
5241:(3): 358–361.
5230:
5207:10.1086/302680
5185:
5149:
5126:10.1086/323299
5119:(3): 615–628.
5104:
5069:
5041:
5006:
4983:10.1086/318205
4976:(2): 432–443.
4960:
4958:
4955:
4954:
4953:
4932:
4877:
4874:
4872:
4871:
4868:on 2011-07-26.
4852:
4837:
4788:
4731:
4703:
4662:
4619:
4596:10.1086/324070
4570:
4513:
4500:
4491:
4438:
4426:
4412:
4402:
4395:South Carolina
4378:
4365:
4336:(3): 454–459.
4315:
4286:(3): 454–459.
4265:
4212:
4159:
4121:
4046:
4037:
4018:News article:
3966:
3943:10.1086/338306
3910:
3887:10.1086/382286
3854:
3802:
3755:
3732:10.1086/340257
3695:
3633:
3580:
3531:
3439:
3394:BMC Evol. Biol
3377:
3333:
3315:
3261:
3200:
3149:
3041:
2992:
2953:(11): e49170.
2930:
2874:
2843:(9): 2603–13.
2823:
2794:(4): 459–466.
2774:
2735:(11): e49170.
2673:Central Africa
2652:
2638:
2579:
2521:
2472:
2420:
2406:
2404:
2401:
2399:
2398:
2393:
2388:
2383:
2378:
2373:
2368:
2363:
2358:
2353:
2347:
2346:
2345:
2341:
2340:
2335:
2330:
2323:
2320:
2319:
2318:
2317:
2316:
2315:
2314:
2306:
2305:
2304:
2303:
2302:
2301:
2300:
2299:
2298:
2286:
2285:
2284:
2266:
2248:
2247:
2246:
2245:
2244:
2243:
2242:
2197:
2182:
2165:
2164:
2158:
2126:
2123:
2121:
2120:
2112:
2104:
2096:
2088:
2080:
2078:Underhill 2000
2072:
2059:
2053:
2052:
2049:
2046:
2043:
2040:
2037:
2034:
2031:
2028:
2025:
2022:
2019:
2016:
2013:
2010:
2007:
2004:
2001:
1998:
1992:
1991:
1988:
1985:
1982:
1979:
1976:
1973:
1970:
1967:
1964:
1961:
1958:
1955:
1952:
1949:
1946:
1943:
1940:
1937:
1931:
1930:
1927:
1924:
1921:
1918:
1915:
1912:
1909:
1906:
1903:
1900:
1897:
1894:
1891:
1888:
1885:
1882:
1879:
1876:
1870:
1869:
1866:
1863:
1860:
1857:
1854:
1851:
1848:
1845:
1842:
1839:
1836:
1833:
1830:
1827:
1824:
1821:
1818:
1815:
1809:
1808:
1805:
1802:
1799:
1796:
1793:
1790:
1787:
1784:
1781:
1778:
1775:
1772:
1769:
1766:
1763:
1760:
1757:
1754:
1748:
1747:
1744:
1741:
1738:
1735:
1732:
1729:
1726:
1723:
1720:
1717:
1714:
1711:
1708:
1705:
1702:
1699:
1696:
1693:
1687:
1686:
1683:
1680:
1677:
1674:
1671:
1668:
1665:
1662:
1659:
1656:
1653:
1650:
1647:
1644:
1641:
1638:
1635:
1632:
1626:
1625:
1622:
1619:
1616:
1613:
1610:
1607:
1604:
1601:
1598:
1595:
1592:
1589:
1586:
1583:
1580:
1577:
1574:
1571:
1565:
1564:
1561:
1558:
1555:
1552:
1549:
1546:
1543:
1540:
1537:
1534:
1531:
1528:
1525:
1522:
1519:
1516:
1513:
1510:
1504:
1503:
1500:
1497:
1494:
1491:
1488:
1485:
1482:
1479:
1476:
1473:
1470:
1467:
1464:
1461:
1458:
1455:
1452:
1449:
1443:
1442:
1439:
1436:
1433:
1430:
1427:
1424:
1421:
1418:
1415:
1412:
1409:
1406:
1403:
1400:
1397:
1394:
1391:
1388:
1343:Main article:
1340:
1337:
1335:
1332:
1235:
1232:
1199:
1196:
1184:Horn of Africa
1179:
1178:A1b1b1 (A-M28)
1176:
1168:Eastern Africa
1163:
1160:
1138:
1137:A1b1a1a (A-M6)
1135:
1071:
1068:
1054:) at 8.3% and
1048:Bakola Pygmies
1040:
1037:
1022:Yemba-speaking
971:
968:
966:
963:
934:
931:
926:
923:
906:
903:
886:
885:Horn of Africa
883:
874:
871:
869:
866:
814:) and eastern
799:
798:Central Africa
796:
792:haplogroup A1a
771:
768:
748:
745:
740:
737:
735:
732:
727:
724:
706:
705:
702:
696:
693:
692:
689:
683:
680:
679:
676:
670:
667:
666:
663:
657:
654:
653:
650:
644:
641:
640:
637:
631:
628:
627:
624:
618:
615:
614:
611:
605:
602:
601:
598:
595:
592:
591:
588:
582:
579:
578:
575:
570:
567:
566:
563:
557:
554:
553:
550:
544:
541:
540:
537:
531:
528:
527:
524:
518:
515:
514:
511:
505:
502:
501:
498:
492:
489:
488:
485:
479:
476:
475:
472:
470:Ethiopian Jews
467:
464:
463:
460:
455:
452:
451:
448:
442:
439:
438:
435:
429:
426:
425:
422:
416:
413:
412:
409:
403:
400:
399:
396:
393:
390:
389:
380:
377:
374:
373:
301:Eastern Africa
262:
259:
243:haplogroup A00
188:Main article:
185:
182:
148:(or among the
133:
132:
96:
92:
91:
85:
81:
80:
78:Central Africa
71:
67:
66:
63:
59:
58:
55:
51:
50:
42:
41:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
6078:
6067:
6064:
6063:
6061:
6039:
6036:
6030:
6027:
6023:
6021:
6016:
6014:
6009:
6002:
5999:
5993:
5990:
5984:
5981:
5975:
5972:
5968:
5964:
5958:
5955:
5949:
5946:
5940:
5937:
5931:
5928:
5922:
5919:
5915:
5914:
5907:
5904:
5899:
5895:
5891:
5887:
5883:
5879:
5876:(2): 187–91.
5875:
5871:
5864:
5861:
5856:
5848:
5845:
5843:
5840:
5839:
5835:
5827:
5825:
5820:
5814:
5811:
5804:
5797:
5792:
5787:
5781:
5775:
5769:
5767:
5765:
5754:
5745:
5738:
5733:
5723:
5714:
5707:
5703:
5697:
5695:
5688:
5681:
5674:
5665:
5659:
5658:
5650:
5645:
5644:
5636:
5631:
5630:
5624:
5619:
5611:
5602:
5597:
5596:
5590:
5587:
5582:
5576:
5573:
5568:
5562:
5559:
5554:
5548:
5545:
5538:
5529:
5521:
5514:
5511:
5506:
5499:
5494:
5488:
5485:
5480:
5474:
5471:
5466:
5460:
5457:
5451:
5444:
5441:
5435:
5428:
5424:
5418:
5411:
5407:
5401:
5394:
5390:
5383:
5378:
5368:
5365:February 2021
5356:
5351:
5342:
5341:
5336:
5332:
5324:
5319:
5317:
5312:
5310:
5305:
5304:
5301:
5295:
5294:
5289:
5286:
5284:
5281:
5279:
5276:
5275:
5271:
5264:
5260:
5256:
5252:
5248:
5247:10.1038/81685
5244:
5240:
5236:
5231:
5227:
5223:
5218:
5213:
5208:
5203:
5199:
5195:
5191:
5186:
5183:
5179:
5175:
5171:
5167:
5163:
5159:
5155:
5150:
5146:
5142:
5137:
5132:
5127:
5122:
5118:
5114:
5110:
5105:
5101:
5097:
5092:
5087:
5084:(2): 97–104.
5083:
5079:
5075:
5070:
5067:
5063:
5059:
5055:
5052:(8): 356–62,
5051:
5047:
5042:
5038:
5034:
5029:
5024:
5020:
5016:
5012:
5007:
5003:
4999:
4994:
4989:
4984:
4979:
4975:
4971:
4967:
4962:
4961:
4956:
4941:
4937:
4933:
4931:
4927:
4924:
4919:
4915:
4910:
4905:
4901:
4897:
4893:
4889:
4885:
4880:
4879:
4875:
4867:
4863:
4856:
4853:
4849:
4848:
4845:ISOGG, 2016,
4841:
4838:
4833:
4829:
4824:
4819:
4815:
4811:
4807:
4803:
4799:
4792:
4789:
4784:
4780:
4775:
4770:
4766:
4762:
4758:
4754:
4750:
4746:
4742:
4735:
4732:
4721:on 2022-08-25
4720:
4716:
4715:
4707:
4704:
4699:
4695:
4690:
4685:
4682:(9): 435–41.
4681:
4677:
4676:J. Hum. Genet
4673:
4666:
4663:
4658:
4654:
4650:
4646:
4642:
4638:
4634:
4630:
4623:
4620:
4615:
4611:
4606:
4601:
4597:
4593:
4589:
4585:
4581:
4574:
4571:
4566:
4560:
4552:
4548:
4544:
4540:
4536:
4532:
4529:(2): 127–48.
4528:
4524:
4517:
4514:
4510:
4504:
4501:
4495:
4492:
4487:
4483:
4479:
4475:
4470:
4469:10400.13/3047
4465:
4461:
4457:
4454:(6): 467–72.
4453:
4449:
4442:
4439:
4435:
4430:
4427:
4422:
4416:
4413:
4406:
4403:
4400:
4396:
4392:
4388:
4382:
4379:
4375:
4369:
4366:
4361:
4357:
4352:
4347:
4343:
4339:
4335:
4331:
4327:
4319:
4316:
4311:
4307:
4302:
4297:
4293:
4289:
4285:
4281:
4277:
4269:
4266:
4261:
4257:
4253:
4249:
4244:
4239:
4235:
4231:
4227:
4223:
4216:
4213:
4208:
4204:
4200:
4196:
4191:
4190:10400.13/3018
4186:
4182:
4178:
4174:
4170:
4163:
4160:
4155:
4151:
4147:
4143:
4140:(3): 387–95.
4139:
4135:
4128:
4126:
4122:
4117:
4113:
4109:
4105:
4101:
4097:
4093:
4089:
4084:
4083:10400.13/4486
4079:
4074:
4069:
4065:
4061:
4057:
4050:
4047:
4041:
4038:
4026:
4022:
4014:
4010:
4005:
4000:
3996:
3992:
3989:(3): 288–93.
3988:
3984:
3980:
3973:
3971:
3967:
3962:
3958:
3953:
3948:
3944:
3940:
3936:
3932:
3928:
3921:
3919:
3917:
3915:
3911:
3906:
3902:
3897:
3892:
3888:
3884:
3881:(3): 532–44.
3880:
3876:
3872:
3865:
3863:
3861:
3859:
3855:
3850:
3846:
3841:
3836:
3832:
3828:
3824:
3820:
3816:
3809:
3807:
3803:
3798:
3794:
3790:
3786:
3782:
3778:
3775:(3): 248–60.
3774:
3770:
3762:
3760:
3756:
3751:
3747:
3742:
3737:
3733:
3729:
3725:
3721:
3717:
3710:
3708:
3706:
3704:
3702:
3700:
3696:
3691:
3687:
3683:
3679:
3675:
3674:10.1038/81685
3671:
3668:(3): 358–61.
3667:
3663:
3656:
3654:
3652:
3650:
3648:
3646:
3644:
3642:
3640:
3638:
3634:
3629:
3625:
3621:
3617:
3614:(3): 316–23.
3613:
3609:
3601:
3599:
3597:
3595:
3593:
3591:
3589:
3587:
3585:
3581:
3576:
3572:
3568:
3564:
3559:
3554:
3550:
3546:
3542:
3535:
3532:
3527:
3523:
3518:
3513:
3510:(7): 867–76.
3509:
3505:
3501:
3494:
3492:
3490:
3488:
3486:
3484:
3482:
3480:
3478:
3476:
3474:
3472:
3470:
3468:
3466:
3464:
3462:
3460:
3458:
3456:
3454:
3452:
3450:
3448:
3446:
3444:
3440:
3435:
3431:
3426:
3421:
3416:
3411:
3407:
3403:
3399:
3395:
3391:
3384:
3382:
3378:
3373:
3369:
3364:
3359:
3356:(7): 1581–9.
3355:
3351:
3347:
3340:
3338:
3334:
3329:
3322:
3320:
3316:
3311:
3307:
3302:
3297:
3292:
3287:
3283:
3279:
3275:
3268:
3266:
3262:
3257:
3253:
3248:
3243:
3238:
3233:
3229:
3225:
3221:
3217:
3213:
3207:
3205:
3201:
3196:
3192:
3187:
3182:
3178:
3174:
3170:
3166:
3162:
3156:
3154:
3150:
3144:
3140:
3135:
3130:
3126:
3122:
3118:
3114:
3110:
3106:
3102:
3095:
3091:
3086:
3081:
3077:
3073:
3069:
3065:
3061:
3057:
3053:
3045:
3042:
3037:
3033:
3028:
3023:
3019:
3015:
3011:
3007:
3003:
2996:
2993:
2988:
2984:
2979:
2974:
2969:
2964:
2960:
2956:
2952:
2948:
2944:
2937:
2935:
2931:
2926:
2922:
2917:
2912:
2908:
2904:
2900:
2896:
2892:
2885:
2883:
2881:
2879:
2875:
2870:
2866:
2861:
2860:10400.13/4486
2856:
2851:
2846:
2842:
2838:
2834:
2827:
2824:
2819:
2815:
2810:
2805:
2801:
2797:
2793:
2789:
2785:
2778:
2775:
2770:
2766:
2761:
2756:
2751:
2746:
2742:
2738:
2734:
2730:
2726:
2720:
2714:
2710:
2705:
2700:
2696:
2692:
2688:
2684:
2680:
2674:
2670:
2666:
2662:
2656:
2653:
2648:
2642:
2639:
2634:
2630:
2625:
2620:
2615:
2610:
2606:
2602:
2598:
2594:
2590:
2583:
2580:
2575:
2571:
2566:
2561:
2557:
2553:
2549:
2545:
2541:
2537:
2533:
2525:
2522:
2517:
2513:
2508:
2503:
2499:
2495:
2492:(4): 728–34.
2491:
2487:
2483:
2476:
2473:
2467:
2463:
2458:
2453:
2449:
2445:
2442:(4): 459–66.
2441:
2437:
2436:
2431:
2424:
2421:
2417:
2411:
2408:
2402:
2397:
2394:
2392:
2389:
2387:
2384:
2382:
2379:
2377:
2374:
2372:
2369:
2367:
2364:
2362:
2359:
2357:
2354:
2352:
2349:
2348:
2343:
2342:
2339:
2336:
2334:
2331:
2329:
2326:
2325:
2321:
2312:
2311:
2307:
2296:
2295:
2294:
2293:formerly A3b2
2291:(M13/PF1374)
2290:
2287:
2282:
2281:
2280:
2279:formerly A3b1
2277:(M51/Page42)
2276:
2273:
2272:
2270:
2267:
2265:
2261:
2258:
2257:
2256:
2252:
2249:
2241:
2237:
2234:
2233:
2231:
2228:
2227:
2226:
2222:
2219:
2218:
2216:
2213:
2212:
2211:(L419/PF712)
2210:
2207:
2206:
2205:
2204:formerly A2-T
2201:
2198:
2195:
2192:
2191:
2190:
2186:
2183:
2181:
2177:
2174:
2173:
2172:
2170:
2162:
2159:
2156:
2153:
2152:
2151:
2149:
2144:
2143:
2138:
2136:
2132:
2124:
2119:
2116:
2113:
2111:
2108:
2105:
2103:
2100:
2097:
2095:
2092:
2089:
2087:
2084:
2081:
2079:
2076:
2073:
2071:
2067:
2064:
2061:
2060:
2058:
2050:
2047:
2044:
2041:
2038:
2035:
2032:
2029:
2026:
2023:
2020:
2017:
2014:
2011:
2008:
2005:
2002:
1999:
1997:
1994:
1993:
1989:
1986:
1983:
1980:
1977:
1974:
1971:
1968:
1965:
1962:
1959:
1956:
1953:
1950:
1947:
1944:
1941:
1938:
1936:
1933:
1932:
1928:
1925:
1922:
1919:
1916:
1913:
1910:
1907:
1904:
1901:
1898:
1895:
1892:
1889:
1886:
1883:
1880:
1877:
1875:
1872:
1871:
1867:
1864:
1861:
1858:
1855:
1852:
1849:
1846:
1843:
1840:
1837:
1834:
1831:
1828:
1825:
1822:
1819:
1816:
1814:
1811:
1810:
1806:
1803:
1800:
1797:
1794:
1791:
1788:
1785:
1782:
1779:
1776:
1773:
1770:
1767:
1764:
1761:
1758:
1755:
1753:
1750:
1749:
1745:
1742:
1739:
1736:
1733:
1730:
1727:
1724:
1721:
1718:
1715:
1712:
1709:
1706:
1703:
1700:
1697:
1694:
1692:
1689:
1688:
1684:
1681:
1678:
1675:
1672:
1669:
1666:
1663:
1660:
1657:
1654:
1651:
1648:
1645:
1642:
1639:
1636:
1633:
1631:
1628:
1627:
1623:
1620:
1617:
1614:
1611:
1608:
1605:
1602:
1599:
1596:
1593:
1590:
1587:
1584:
1581:
1578:
1575:
1572:
1570:
1567:
1566:
1562:
1559:
1556:
1553:
1550:
1547:
1544:
1541:
1538:
1535:
1532:
1529:
1526:
1523:
1520:
1517:
1514:
1511:
1509:
1506:
1505:
1501:
1498:
1495:
1492:
1489:
1486:
1483:
1480:
1477:
1474:
1471:
1468:
1465:
1462:
1459:
1456:
1453:
1450:
1448:
1445:
1444:
1440:
1437:
1434:
1431:
1428:
1425:
1422:
1419:
1416:
1413:
1410:
1407:
1404:
1401:
1398:
1395:
1392:
1389:
1386:
1385:
1382:
1379:
1375:
1374:Cruciani 2011
1370:
1368:
1364:
1363:Haplogroup BT
1355:
1351:
1346:
1338:
1334:Phylogenetics
1333:
1331:
1324:
1320:
1317:
1315:
1311:
1307:
1303:
1299:
1295:
1292:(1/20 = 5%),
1291:
1290:Yemenite Jews
1287:
1283:
1282:Aegean Region
1278:
1276:
1272:
1268:
1263:
1261:
1257:
1253:
1249:
1245:
1240:
1233:
1231:
1229:
1225:
1221:
1217:
1213:
1212:Bantu peoples
1209:
1205:
1197:
1195:
1193:
1189:
1185:
1177:
1175:
1173:
1169:
1162:A1b1b (A-M32)
1161:
1159:
1157:
1153:
1149:
1145:
1136:
1134:
1131:
1128:
1124:
1119:
1117:
1113:
1109:
1105:
1101:
1097:
1093:
1089:
1085:
1081:
1077:
1076:Guinea-Bissau
1069:
1067:
1065:
1061:
1057:
1053:
1049:
1044:
1038:
1036:
1034:
1030:
1025:
1023:
1020:(or Nweh), a
1017:
1012:
1007:
1005:
1001:
997:
992:
990:
986:
982:
978:
970:A00 (A00-AF6)
969:
964:
962:
960:
959:haplogroup CT
956:
955:haplogroup BT
952:
948:
944:
940:
932:
930:
924:
922:
920:
916:
912:
904:
902:
900:
896:
892:
884:
882:
880:
872:
867:
865:
863:
859:
855:
851:
847:
843:
838:
836:
831:
829:
825:
821:
817:
813:
810:, 2/17 = 12%
809:
806:, 4/28 = 14%
805:
797:
795:
793:
789:
785:
781:
777:
776:Guinea Bissau
769:
767:
765:
760:
758:
754:
746:
744:
738:
733:
731:
726:North America
725:
723:
722:from Sudan.
721:
720:Nilo-Saharans
717:
713:
703:
700:
697:
695:
694:
690:
687:
686:South Semitic
684:
682:
681:
677:
674:
671:
669:
668:
664:
661:
658:
656:
655:
651:
648:
645:
643:
642:
638:
635:
632:
630:
629:
625:
622:
619:
617:
616:
612:
609:
606:
604:
603:
599:
596:
594:
593:
589:
586:
583:
581:
580:
576:
574:
571:
569:
568:
564:
561:
558:
556:
555:
551:
548:
545:
543:
542:
538:
535:
532:
530:
529:
525:
522:
519:
517:
516:
512:
509:
506:
504:
503:
499:
496:
493:
491:
490:
486:
483:
480:
478:
477:
473:
471:
468:
466:
465:
461:
459:
456:
454:
453:
449:
446:
443:
441:
440:
436:
433:
430:
428:
427:
423:
420:
417:
415:
414:
410:
407:
404:
402:
401:
397:
394:
392:
391:
384:
381:
378:
376:
375:
372:
368:
365:
360:
358:
354:
350:
346:
342:
338:
334:
330:
326:
322:
317:
314:
310:
306:
302:
298:
294:
290:
287:
282:
280:
276:
272:
268:
260:
258:
256:
252:
251:KwaZulu-Natal
247:
244:
239:
236:
232:
227:
225:
221:
217:
216:haplogroup BT
213:
204:
196:
191:
183:
181:
179:
175:
171:
167:
163:
159:
155:
154:haplogroup BT
151:
147:
143:
139:
130:
126:
122:
118:
114:
110:
105:
102:(AF6/L1284),
101:
97:
93:
89:
86:
82:
79:
75:
72:
68:
64:
60:
56:
52:
48:
43:
38:
33:
19:
6038:
6029:
6019:
6012:
6007:
6001:
5992:
5983:
5974:
5957:
5948:
5939:
5930:
5921:
5912:
5906:
5873:
5869:
5863:
5854:
5719:
5662:
5362:
5354:
5291:
5238:
5234:
5197:
5193:
5157:
5153:
5116:
5112:
5081:
5077:
5049:
5045:
5018:
5014:
4973:
4969:
4944:. Retrieved
4940:the original
4894:(3): 454–9.
4891:
4887:
4876:Bibliography
4866:the original
4855:
4846:
4840:
4808:(5): 830–8.
4805:
4801:
4791:
4748:
4744:
4734:
4723:. Retrieved
4719:the original
4713:
4706:
4679:
4675:
4665:
4632:
4628:
4622:
4587:
4583:
4573:
4559:cite journal
4526:
4522:
4516:
4508:
4503:
4494:
4451:
4447:
4441:
4433:
4429:
4415:
4405:
4381:
4368:
4333:
4329:
4318:
4283:
4279:
4268:
4225:
4221:
4215:
4172:
4168:
4162:
4137:
4133:
4063:
4059:
4049:
4040:
4029:. Retrieved
4027:. 2007-01-24
3986:
3982:
3937:(1): 265–8.
3934:
3930:
3878:
3874:
3825:(7): 800–7.
3822:
3818:
3772:
3768:
3723:
3719:
3665:
3661:
3611:
3607:
3548:
3544:
3534:
3507:
3503:
3397:
3393:
3353:
3349:
3281:
3277:
3219:
3215:
3168:
3164:
3108:
3104:
3059:
3055:
3044:
3009:
3005:
2995:
2950:
2946:
2901:(6): 814–8.
2898:
2894:
2840:
2836:
2826:
2791:
2787:
2777:
2732:
2728:
2718:
2689:(6): 814–8.
2686:
2682:
2660:
2655:
2641:
2596:
2592:
2582:
2539:
2535:
2524:
2489:
2485:
2475:
2439:
2433:
2423:
2416:human Y-MRCA
2410:
2308:
2292:
2288:
2278:
2274:
2268:
2264:formerly A3a
2263:
2259:
2254:
2250:
2239:
2235:
2229:
2224:
2220:
2214:
2208:
2203:
2199:
2193:
2188:
2184:
2180:formerly A1b
2179:
2175:
2168:
2167:
2166:
2160:
2154:
2150:(AF6/L1284)
2147:
2146:
2141:
2140:
2139:
2128:
2118:Capelli 2001
2114:
2106:
2098:
2094:Karafet 2001
2090:
2082:
2074:
2062:
2056:
1371:
1360:
1348:
1329:
1318:
1279:
1264:
1260:Hausa people
1241:
1237:
1220:Sotho–Tswana
1201:
1181:
1165:
1140:
1132:
1120:
1083:
1073:
1045:
1042:
1026:
1008:
1003:
993:
988:
981:Jebel Irhoud
973:
957:, including
936:
928:
921:population.
908:
888:
876:
839:
832:
826:, 1/47 = 2%
822:, 1/18 = 6%
801:
773:
766:males (3%).
761:
750:
742:
739:North Africa
729:
709:
370:
363:
337:Haplogroup E
318:
283:
279:Western Asia
264:
261:Distribution
255:South Africa
248:
240:
228:
209:
177:
173:
169:
165:
138:Haplogroup A
137:
136:
88:Human Y-MRCA
40:Haplogroup A
4751:(1): 6468.
4434:Lipson Mark
4391:York County
2647:"A00 YTree"
2255:formerly A3
2225:formerly A2
2102:Semino 2000
2086:Hammer 2001
1441:ISOGG 2012
1438:ISOGG 2011
1435:ISOGG 2010
1432:ISOGG 2009
1429:ISOGG 2008
1426:ISOGG 2007
1423:ISOGG 2006
1070:A1a (A-M31)
1039:A0 (A-V148)
1014: [
1006:(A-A4987).
991:(A-L1149).
891:Beta Israel
868:East Africa
818:(2/9 = 22%
770:West Africa
716:Beta Israel
688:(Ethiopia)
662:(Ethiopia)
636:(Cameroon)
387:(in %)
341:West Africa
329:Niger–Congo
95:Descendants
4862:"YCC Tree"
4725:2022-08-25
4523:Hum. Genet
4448:Hum. Genet
4243:2108/37090
4031:2007-01-27
3769:Hum. Mutat
3662:Nat. Genet
3400:(1): 124.
2403:References
1685:A1b1a1a1b
1624:A1b1a1a1a
1096:Senegambia
1088:Cabo Verde
979:, such as
788:Cabo Verde
747:Upper Nile
675:(Eritrea)
649:(Namibia)
573:Ethiopians
549:(Eritrea)
408:(Namibia)
299:groups in
231:San people
6020:PhyloTree
5855:Footnotes
4100:748733133
4092:0737-4038
3278:BMC Genet
2717:Scozzari
2669:Northwest
2051:A1b1b2b1
1378:insertion
1123:Yorkshire
1029:Shum Laka
998:males of
965:Subclades
919:Khoisanid
98:primary:
6060:Category
6017:, and;
6008:op. cit.
5898:23291764
5890:24166809
5528:F-Y27277
5263:12893406
5255:11062480
5226:10577926
5182:11073453
5145:11481588
5100:11313742
5066:10904265
5037:11420360
5002:11170891
4946:30 March
4926:Archived
4918:23453668
4832:18385274
4783:35618734
4698:16142507
4657:11073453
4614:11573163
4551:10763736
4543:14586639
4486:63381583
4478:12942365
4360:23453668
4310:23453668
4260:25536759
4252:16626331
4199:15996172
4154:12927125
4116:11190055
4108:21478374
4025:BBC News
4013:17245408
3961:11719903
3905:14973781
3849:20051990
3789:15300852
3750:11910562
3690:12893406
3682:11062480
3628:18618658
3567:11415522
3526:15856073
3434:17662131
3372:19369595
3310:19772609
3256:26226630
3216:PLOS ONE
3195:25770088
3143:23908239
3094:23908240
3036:28971970
2987:23145109
2947:PLOS ONE
2925:21601174
2869:21478374
2818:25770088
2769:23145109
2729:PLOS ONE
2713:21601174
2633:26226630
2593:PLOS ONE
2574:31969706
2516:27058445
2466:25770088
2322:See also
2236:A1b1a1a1
2171:(L1085)
1990:removed
1929:A1b1b2b
1868:A1b1b2a
1563:A1b1a1a
1302:Sardinia
1267:Ethiopia
1092:Mandinka
1052:Cameroon
951:Cypriots
939:Mitilini
790:carried
764:Egyptian
757:Sudanese
701:(Egypt)
610:(Sudan)
587:(Kenya)
562:(Sudan)
536:(Kenya)
523:(Sudan)
510:(Sudan)
497:(Sudan)
484:/Sekele
447:(Sudan)
434:(Sudan)
421:(Sudan)
321:Ethiopia
84:Ancestor
5732:K-M2313
5725:
5666:
5651:
5637:
5621:
5612:
5603:
5425:
5408:
5355:updated
5290:, from
5217:1288383
5162:Bibcode
5154:Science
5136:1235490
4993:1235276
4909:3591855
4823:2336805
4774:9135728
4753:Bibcode
4637:Bibcode
4629:Science
4605:1274378
4351:3591855
4301:3591855
4207:3229760
4004:2590664
3896:1182266
3840:2987365
3797:1571356
3575:9441236
3425:1976131
3402:Bibcode
3301:2759955
3247:4520482
3224:Bibcode
3186:4381518
3134:4032117
3113:Bibcode
3105:Science
3085:5500864
3064:Bibcode
3056:Science
3014:Bibcode
3006:Science
2978:3492319
2955:Bibcode
2916:3113241
2809:4381518
2760:3492319
2737:Bibcode
2704:3113241
2624:4520482
2601:Bibcode
2565:8386425
2544:Bibcode
2507:4833433
2457:4381518
2289:A1b1b2b
2275:A1b1b2a
2271:(L427)
2230:A1b1a1a
2202:(P108)
2187:(P305)
2110:Su 1999
1807:A1b1b1
1192:Somalis
1127:England
1112:Morocco
1108:Berbers
1060:Algeria
1056:Berbers
1050:(South
947:Madeira
915:Hadzabe
911:Khoisan
808:Mandara
712:Khoisan
560:Masalit
458:Khoisan
432:Shilluk
357:Khoisan
335:clade.
305:Central
297:Nilotic
286:Bushmen
90:(A00-T)
5967:K-M526
5896:
5888:
5755:
5751:
5746:
5734:
5715:
5542:
5534:
5530:
5525:
5261:
5253:
5224:
5214:
5180:
5143:
5133:
5098:
5064:
5035:
5000:
4990:
4923:as PDF
4916:
4906:
4830:
4820:
4781:
4771:
4696:
4655:
4612:
4602:
4549:
4541:
4509:et al.
4484:
4476:
4397:. See
4374:Y-MRCA
4358:
4348:
4308:
4298:
4258:
4250:
4205:
4197:
4152:
4114:
4106:
4098:
4090:
4011:
4001:
3959:
3952:384897
3949:
3903:
3893:
3847:
3837:
3795:
3787:
3748:
3741:447595
3738:
3688:
3680:
3626:
3573:
3565:
3524:
3432:
3422:
3370:
3308:
3298:
3284:: 59.
3254:
3244:
3193:
3183:
3141:
3131:
3092:
3082:
3034:
2985:
2975:
2923:
2913:
2867:
2816:
2806:
2767:
2757:
2719:et al.
2711:
2701:
2661:et al.
2631:
2621:
2572:
2562:
2536:Nature
2514:
2504:
2464:
2454:
2366:A-M171
2356:A-M118
2351:A-M114
2269:A1b1b2
2262:(M28)
2260:A1b1b1
2253:(M32)
2238:(P28)
2223:(M14)
2221:A1b1a1
1996:A-M118
1935:A-M171
1746:A1b1b
1569:A-M114
1306:Jordan
1286:Turkey
1271:Amhara
1084:et al.
1011:Bangwa
943:Lesvos
933:Europe
895:Amhara
835:Bakola
804:Tupuri
734:Africa
718:, and
673:Kunama
534:Maasai
371:Africa
275:Europe
271:Africa
190:Y-MRCA
184:Origin
176:, and
146:Africa
5894:S2CID
5544:GHIJK
5259:S2CID
4547:S2CID
4482:S2CID
4387:slave
4256:S2CID
4203:S2CID
4112:S2CID
3793:S2CID
3686:S2CID
3571:S2CID
2396:A-P28
2386:A-M51
2381:A-M32
2376:A-M31
2371:A-M28
2361:A-M13
2313:(M91)
2251:A1b1b
2232:(M6)
2215:A1b1a
2196:(M31)
2131:ISOGG
2048:A3b2b
2045:A3b2b
2042:A3b2b
2039:A3b2b
2036:A3b2b
2033:A3b2b
2030:A3b2b
2027:A3b2b
2024:A3b2b
2021:A3b2b
1987:A3b2a
1984:A3b2a
1981:A3b2a
1978:A3b2a
1975:A3b2a
1972:A3b2a
1969:A3b2a
1966:A3b2a
1963:A3b2a
1960:A3b2a
1899:A3b2*
1874:A-M13
1813:A-M51
1752:A-M28
1691:A-M32
1630:A-P28
1447:A-M31
1365:from
1310:Amman
1294:Egypt
1275:Oromo
1244:Sudan
1224:Xhosa
1208:!Kung
1148:!Kung
1144:!Kung
1110:from
1102:from
1100:Dogon
1094:from
1064:Ghana
1058:from
1033:Mbuti
1018:]
899:Oromo
879:Iraqw
862:Tsogo
858:Nzebi
850:Ndumu
842:Gabon
828:Mbuti
812:Fulbe
780:Niger
699:Arabs
660:Oromo
634:Fulbe
608:Hausa
585:Bantu
495:Borgu
482:!Kung
419:Dinka
383:Freq.
353:Mbuti
349:B-M60
333:E1b1a
325:Pygmy
140:is a
5886:PMID
5713:K2b1
5558:HIJK
5450:A1b1
5406:A0-T
5251:PMID
5222:PMID
5178:PMID
5141:PMID
5096:PMID
5062:PMID
5033:PMID
4998:PMID
4948:2013
4914:PMID
4828:PMID
4779:PMID
4694:PMID
4653:PMID
4610:PMID
4565:link
4539:PMID
4474:PMID
4356:PMID
4306:PMID
4248:PMID
4195:PMID
4150:PMID
4104:PMID
4096:OCLC
4088:ISSN
4009:PMID
3957:PMID
3901:PMID
3845:PMID
3785:PMID
3746:PMID
3678:PMID
3624:PMID
3563:PMID
3522:PMID
3430:PMID
3368:PMID
3306:PMID
3252:PMID
3191:PMID
3139:PMID
3090:PMID
3032:PMID
2983:PMID
2921:PMID
2865:PMID
2814:PMID
2765:PMID
2709:PMID
2671:and
2665:West
2629:PMID
2570:PMID
2512:PMID
2462:PMID
2391:A-M6
2209:A1b1
2169:A0-T
2161:A00b
2155:A00a
2068:and
1926:A3b2
1923:A3b2
1920:A3b2
1917:A3b2
1914:A3b2
1911:A3b2
1908:A3b2
1905:A3b2
1902:A3b2
1865:A3b1
1862:A3b1
1859:A3b1
1856:A3b1
1853:A3b1
1850:A3b1
1847:A3b1
1844:A3b1
1841:A3b1
1838:A3b1
1508:A-M6
1502:A1a
1408:(η)
1405:(ζ)
1402:(ε)
1399:(δ)
1396:(γ)
1393:(β)
1390:(α)
1314:Oman
1252:Nuba
1228:Zulu
1204:Nama
1170:and
1156:Dama
1152:Nama
1116:Mali
1104:Mali
1004:A00b
989:A00a
925:Asia
854:Duma
846:Baka
824:Hema
820:Alur
784:Mali
647:Dama
621:Khwe
547:Nara
508:Nuer
487:~40
445:Nuba
406:Nama
398:66%
277:and
178:A1b1
127:and
125:A1b1
104:A0-T
5878:doi
5796:P1a
5791:P1b
5786:P1c
5774:NO1
5702:K2a
5694:K2b
5687:K2c
5680:K2d
5673:K2e
5567:IJK
5440:A1b
5434:A1a
5400:A00
5243:doi
5212:PMC
5202:doi
5170:doi
5158:290
5131:PMC
5121:doi
5086:doi
5054:doi
5023:doi
4988:PMC
4978:doi
4904:PMC
4896:doi
4818:PMC
4810:doi
4769:PMC
4761:doi
4684:doi
4645:doi
4633:290
4600:PMC
4592:doi
4531:doi
4527:114
4464:hdl
4456:doi
4452:113
4346:PMC
4338:doi
4296:PMC
4288:doi
4238:hdl
4230:doi
4185:hdl
4177:doi
4142:doi
4078:hdl
4068:doi
3999:PMC
3991:doi
3947:PMC
3939:doi
3891:PMC
3883:doi
3835:PMC
3827:doi
3777:doi
3736:PMC
3728:doi
3670:doi
3616:doi
3612:137
3553:doi
3512:doi
3420:PMC
3410:doi
3358:doi
3296:PMC
3286:doi
3242:PMC
3232:doi
3181:PMC
3173:doi
3129:PMC
3121:doi
3109:341
3080:PMC
3072:doi
3060:341
3022:doi
3010:358
2973:PMC
2963:doi
2911:PMC
2903:doi
2855:hdl
2845:doi
2804:PMC
2796:doi
2755:PMC
2745:doi
2699:PMC
2691:doi
2619:PMC
2609:doi
2560:PMC
2552:doi
2540:577
2502:PMC
2494:doi
2452:PMC
2444:doi
2200:A1b
2194:A1a
2148:A00
2135:YCC
2012:Eu1
1951:Eu1
1890:Eu1
1804:A3a
1801:A3a
1798:A3a
1795:A3a
1792:A3a
1789:A3a
1786:A3a
1783:A3a
1780:A3a
1777:A3a
1682:A2b
1679:A2b
1676:A2b
1673:A2b
1670:A2b
1667:A2b
1664:A2b
1661:A2b
1658:A2b
1655:A2b
1621:A2a
1618:A2a
1615:A2a
1612:A2a
1609:A2a
1606:A2a
1603:A2a
1600:A2a
1597:A2a
1594:A2a
1533:A2*
1499:A1a
1496:A1a
1493:A1a
1490:A1a
1481:A1a
1284:of
1265:In
1242:In
1214:of
996:Mbo
830:).
816:DRC
691:10
678:10
665:10
652:11
639:12
626:12
613:13
600:14
590:14
577:14
565:19
552:20
539:27
526:31
521:Fur
513:33
500:35
474:41
462:44
450:46
437:53
424:62
411:64
224:kya
174:A1a
166:A00
160:of
121:A1b
117:A1a
100:A00
6062::
6010:;
5892:.
5884:.
5874:35
5872:.
5764:P1
5649:J2
5643:J1
5635:I2
5629:I1
5618:K2
5610:LT
5581:IJ
5537:F3
5520:F1
5484:CF
5479:DE
5470:CT
5456:BT
5423:A1
5417:A0
5391:"
5257:.
5249:.
5239:26
5237:.
5220:.
5210:.
5198:65
5196:.
5192:.
5176:,
5168:,
5156:,
5139:.
5129:.
5117:69
5115:.
5111:.
5094:.
5080:.
5076:.
5060:,
5050:16
5048:,
5031:.
5019:18
5017:.
5013:.
4996:.
4986:.
4974:68
4972:.
4968:.
4912:.
4902:.
4892:92
4890:.
4886:.
4826:.
4816:.
4806:18
4804:.
4800:.
4777:.
4767:.
4759:.
4749:12
4747:.
4743:.
4692:.
4680:50
4678:.
4674:.
4651:.
4643:.
4631:.
4608:.
4598:.
4588:69
4586:.
4582:.
4561:}}
4557:{{
4545:.
4537:.
4525:.
4480:.
4472:.
4462:.
4450:.
4393:,
4354:.
4344:.
4334:92
4332:.
4328:.
4304:.
4294:.
4284:92
4282:.
4278:.
4254:.
4246:.
4236:.
4226:70
4224:.
4201:.
4193:.
4183:.
4173:69
4171:.
4148:.
4138:28
4136:.
4124:^
4110:.
4102:.
4094:.
4086:.
4076:.
4064:28
4062:.
4058:.
4023:.
4007:.
3997:.
3987:15
3985:.
3981:.
3969:^
3955:.
3945:.
3935:70
3933:.
3929:.
3913:^
3899:.
3889:.
3879:74
3877:.
3873:.
3857:^
3843:.
3833:.
3823:18
3821:.
3817:.
3805:^
3791:.
3783:.
3773:24
3771:.
3758:^
3744:.
3734:.
3724:70
3722:.
3718:.
3698:^
3684:.
3676:.
3666:26
3664:.
3636:^
3622:.
3610:.
3583:^
3569:.
3561:.
3549:65
3547:.
3543:.
3520:.
3508:13
3506:.
3502:.
3442:^
3428:.
3418:.
3408:.
3396:.
3392:.
3380:^
3366:.
3354:26
3352:.
3348:.
3336:^
3318:^
3304:.
3294:.
3282:10
3280:.
3276:.
3264:^
3250:.
3240:.
3230:.
3220:10
3218:.
3214:.
3203:^
3189:.
3179:.
3169:25
3167:.
3163:.
3152:^
3137:.
3127:.
3119:.
3107:.
3103:.
3088:.
3078:.
3070:.
3058:.
3054:.
3030:.
3020:.
3008:.
3004:.
2981:.
2971:.
2961:.
2949:.
2945:.
2933:^
2919:.
2909:.
2899:88
2897:.
2893:.
2877:^
2863:.
2853:.
2841:28
2839:.
2835:.
2812:.
2802:.
2792:25
2790:.
2786:.
2763:.
2753:.
2743:.
2731:.
2727:.
2707:.
2697:.
2687:88
2685:.
2681:.
2667:,
2627:.
2617:.
2607:.
2597:10
2595:.
2591:.
2568:.
2558:.
2550:.
2538:.
2534:.
2510:.
2500:.
2490:98
2488:.
2484:.
2460:.
2450:.
2440:25
2438:.
2432:.
2310:BT
2185:A1
2176:A0
2133:,
2015:H1
2006:1A
1954:H1
1945:1A
1893:H1
1884:1A
1832:H1
1823:1A
1771:H1
1762:1A
1743:A3
1740:A3
1737:A3
1734:A3
1731:A3
1728:A3
1725:A3
1722:A3
1719:A3
1649:H1
1634:27
1588:H1
1573:27
1560:A2
1557:A2
1554:A2
1551:A2
1548:A2
1545:A2
1542:A2
1539:A2
1536:A2
1527:H1
1512:27
1487:A1
1484:A1
1478:A1
1475:A1
1472:A1
1466:H1
1457:1A
1308:,
1230:.
1174:.
1125:,
1118:.
1066:.
1016:fr
961:.
864:.
794:.
704:3
359:.
281:.
267:BT
253:,
235:L0
172:,
170:A0
168:,
131:.)
129:BT
123:,
119:,
115:,
113:A1
111:,
109:A0
76:,
5900:.
5880::
5824:Q
5819:R
5810:O
5803:N
5753:M
5744:S
5722:P
5664:T
5657:L
5601:J
5595:I
5586:K
5572:H
5553:G
5510:F
5505:C
5498:E
5493:D
5465:B
5387:"
5367:)
5363:(
5357:.
5322:e
5315:t
5308:v
5265:.
5245::
5228:.
5204::
5172::
5164::
5147:.
5123::
5102:.
5088::
5082:9
5056::
5039:.
5025::
5004:.
4980::
4950:.
4920:.
4898::
4834:.
4812::
4785:.
4763::
4755::
4728:.
4700:.
4686::
4659:.
4647::
4639::
4616:.
4594::
4567:)
4553:.
4533::
4488:.
4466::
4458::
4362:.
4340::
4312:.
4290::
4262:.
4240::
4232::
4209:.
4187::
4179::
4156:.
4144::
4118:.
4080::
4070::
4034:.
4015:.
3993::
3963:.
3941::
3907:.
3885::
3851:.
3829::
3799:.
3779::
3752:.
3730::
3692:.
3672::
3630:.
3618::
3577:.
3555::
3528:.
3514::
3436:.
3412::
3404::
3398:7
3374:.
3360::
3330:.
3312:.
3288::
3258:.
3234::
3226::
3197:.
3175::
3145:.
3123::
3115::
3096:.
3074::
3066::
3038:.
3024::
3016::
2989:.
2965::
2957::
2951:7
2927:.
2905::
2871:.
2857::
2847::
2820:.
2798::
2771:.
2747::
2739::
2733:7
2715:.
2693::
2649:.
2635:.
2611::
2603::
2576:.
2554::
2546::
2518:.
2496::
2468:.
2446::
2115:η
2107:ζ
2099:ε
2091:δ
2083:γ
2075:β
2063:α
2018:A
2009:2
2003:I
2000:7
1957:A
1948:2
1942:I
1939:7
1896:A
1887:2
1881:I
1878:7
1835:A
1829:–
1826:1
1820:I
1817:7
1774:A
1768:–
1765:1
1759:I
1756:7
1716:*
1713:*
1710:*
1707:*
1704:*
1701:*
1698:*
1695:*
1652:A
1646:–
1643:4
1640:2
1637:I
1591:A
1585:–
1582:3
1579:2
1576:I
1530:A
1524:–
1521:3
1518:2
1515:I
1469:A
1463:–
1460:1
1454:I
1451:7
307:-
34:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.