124:, and has not been widely implemented in clinical settings although its impact could be quite large. Although blood-borne ctDNA remains the most clinically significant noninvasive cancer detection, other studies have emerged that investigate other potential methods, including detection of colorectal cancer via fecal samples.
36:
and DNA sampling techniques that often required larger samples of tissue or the destruction of the studied organism. Noninvasive genotyping is widely utilized in conservation efforts, where capture and sampling may be difficult or disruptive to behavior. Additionally, in medicine, this technique is
119:
occurs and, therefore, detection of certain mutant alleles may enhance survival rates in cancer patients. In a recent study, ctDNA was shown to be "a broadly applicable, sensitive, and specific biomarker that can be used for a variety of clinical and research purposes in patients with multiple
49:. Modern DNA amplification methods allow researchers to use a variety of animal material collected in the field, including feces, hair, and feathers, to gain insights into effective population size, gene flow, and hybridization. Despite the potential that noninvasive genotyping has in
132:
The method by which samples are collected in noninvasive genotyping is what separates the technique from traditional genotyping, and there are a number of ways that this is accomplished. In the field, procured samples of tissue are captured, the tissue is dissolved, and the DNA is
53:
efforts, it is still not broadly used, potentially due to problems with degradation, contamination or a lower DNA quality in comparison with blood or tissue samples. However, optimized laboratory protocols and specialized extraction kits can help overcome these issues.
19:
is a modern technique for obtaining DNA for genotyping that is characterized by the indirect sampling of specimen, not requiring harm to, handling of, or even the presence of the organism of interest. Beginning in the early 1990s, with the advent of
37:
being applied in humans for the diagnosis of genetic disease and early detection of tumors. In this context, invasivity takes on a separate definition where noninvasive sampling also includes simple blood samples.
426:
Olah G, Heinsohn RG, Brightsmith DJ, Espinoza JR, Peakall R (December 2016). "Validation of non-invasive genetic tagging in two large macaw species (Ara macao and A. chloropterus) of the
Peruvian Amazon".
87:
as it requires only a simple blood sample. One NIPD provider maintains that a 10 mL blood sample will provide 99% accurate detection of basic genomic abnormalities as early as 10 weeks into pregnancy. The
188:
1003:
Spethmann S, Fischer C, Wagener C, Streichert T, Tschentscher P (March 2004). "Nucleic acids from intact epithelial cells as a target for stool-based molecular diagnosis of colorectal cancer".
137:, although the exact procedure differs between different samples. Following the collection of DNA samples, PCR technology is utilized to amplify particular genetic sequences, with PCR
318:
Powell C, Butler F, O'Reilly C (December 2019). "The development of real-time PCR assays for species and sex identification of three sympatric deer species from noninvasive samples".
111:
This same technique is also utilized to identify the incidence of tumor DNA in the blood, which can both provide early detection of tumor growth and indicate relapse in cancer.
101:
141:
specificity avoiding contamination from other DNA sources. Then, the DNA can be analyzed using a number of genomic techniques, similarly to traditionally obtained samples.
171:
743:"Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma"
974:
100:
189:"Towards more compassionate wildlife research through the 3Rs principles: moving from invasive to non-invasive methods"
46:
138:
21:
371:"Are shed hair genomes the most effective noninvasive resource for estimating relationships in the wild?"
808:
112:
50:
754:
697:
640:
629:"De novo discovery of SNPs for genotyping endangered sun parakeets (Aratinga solstitialis) in Guyana"
491:
436:
382:
327:
274:
76:
772:
666:
460:
351:
228:
68:
1093:
1069:
1020:
955:
906:
853:
834:"Noninvasive Genotyping and Assessment of Treatment Response in Diffuse Large B Cell Lymphoma"
790:
723:
658:
609:
558:
517:
452:
408:
369:
Khan A, Patel K, Bhattacharjee S, Sharma S, Chugani AN, Sivaraman K, et al. (June 2020).
343:
300:
220:
167:
159:
1059:
1051:
1012:
945:
937:
896:
888:
872:
845:
780:
762:
713:
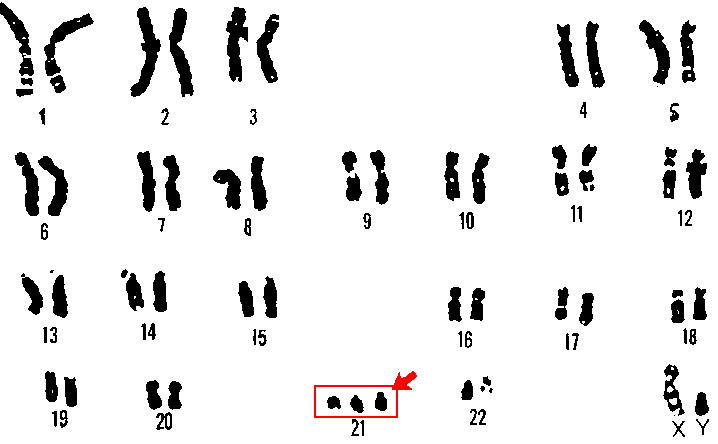
705:
648:
599:
589:
548:
507:
499:
444:
398:
390:
335:
290:
282:
210:
200:
134:
84:
982:
924:
Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. (February 2014).
758:
741:
Lun FM, Tsui NB, Chan KC, Leung TY, Lau TK, Charoenkwan P, et al. (December 2008).
701:
644:
495:
440:
386:
331:
278:
1064:
1039:
950:
925:
901:
876:
785:
742:
718:
685:
604:
578:"From promise to practice: pairing non-invasive sampling with genomics in conservation"
577:
536:
512:
479:
403:
370:
295:
262:
553:
478:
Carroll EL, Bruford MW, DeWoody JA, Leroy G, Strand A, Waits L, Wang J (August 2018).
1087:
670:
464:
355:
286:
232:
121:
93:
72:
1040:"A simple method of genomic DNA extraction from human samples for PCR-RFLP analysis"
832:
Scherer F, Kurtz DM, Newman AM, Stehr H, Liu CL, Zhou L, et al. (2015-12-03).
877:"Blood circulating tumor DNA for non-invasive genotyping of colon cancer patients"
849:
941:
892:
833:
79:
in maternal plasma, NIPD became a popular method for determining sex, paternity,
45:
In conservation, noninvasive genotyping is an important part of implementing the
926:"Detection of circulating tumor DNA in early- and late-stage human malignancies"
29:
747:
Proceedings of the
National Academy of Sciences of the United States of America
653:
628:
448:
339:
116:
80:
857:
662:
562:
456:
347:
224:
160:"Noninvasive genotyping and field studies of free-ranging nonhuman primates."
28:
samples from small quantities of hair, feathers, scales, or excrement. These
1055:
813:
The NIFTY™ Test - A Non-Invasive
Prenatal Test Brought To By BGI Diagnostics
767:
89:
1073:
1024:
959:
910:
794:
727:
613:
537:"Poor implementation of non-invasive sampling in wildlife genetics studies"
521:
412:
304:
1016:
627:
Spitzer R, Norman AJ, Königsson H, Schiffthaler B, Spong G (2020-12-01).
480:"Genetic and genomic monitoring with minimally invasive sampling methods"
33:
776:
709:
684:
Fan HC, Gu W, Wang J, Blumenfeld YJ, El-Sayed YY, Quake SR (July 2012).
215:
594:
503:
394:
205:
120:
different types of cancer". This technique is often referred to as a
71:(NIPD), which provides an alternative to riskier techniques such as
263:"Genomic-scale capture and sequencing of endogenous DNA from feces"
576:
Russello MA, Waterhouse MD, Etter PD, Johnson EA (2015-07-21).
96:, which is what is most routinely checked for by NIPD screens.
25:
67:
The most common use of noninvasive genotyping in medicine is
247:
1038:
Ghatak S, Muthukumaran RB, Nachimuthu SK (December 2013).
261:
Perry GH, Marioni JC, Melsted P, Gilad Y (December 2010).
686:"Non-invasive prenatal measurement of the fetal genome"
24:, researchers have been able to obtain high-quality
92:below is that of an individual with trisomy 21, or
809:"NIFTY test methodology and sequencing technology"
166:. Oxford University Press, USA. pp. 46–68.
8:
1005:International Journal of Molecular Medicine
1063:
949:
900:
784:
766:
717:
652:
603:
593:
552:
511:
402:
294:
248:"The 3Rs Principles in Wildlife Research"
214:
204:
975:"Blood Test for Early Cancer Detection"
150:
32:samples are an improvement over older
7:
883:. Thematic Issue: Liquid biopsies.
1044:Journal of Biomolecular Techniques
162:. In Chapais B, Berman CM (eds.).
14:
554:10.3897/rethinkingecology.4.32751
115:can be found in the blood before
287:10.1111/j.1365-294X.2010.04888.x
164:Kinship and Behavior in Primates
99:
633:Conservation Genetics Resources
429:Conservation Genetics Resources
320:Conservation Genetics Resources
69:non-invasive prenatal diagnosis
930:Science Translational Medicine
1:
850:10.1182/blood.V126.23.114.114
942:10.1126/scitranslmed.3007094
893:10.1016/j.molonc.2015.12.005
1110:
654:10.1007/s12686-020-01151-x
535:Zemanova MA (2019-07-16).
246:Zemanova MA (2023-11-30).
187:Zemanova MA (2020-03-17).
158:Woodruff DS (March 2004).
484:Evolutionary Applications
449:10.1007/s12686-016-0573-4
340:10.1007/s12686-018-1041-0
83:, and the occurrence of
75:. With the discovery of
1056:10.7171/jbt.13-2404-001
768:10.1073/pnas.0810373105
17:Noninvasive genotyping
1017:10.3892/ijmm.13.3.451
979:MIT Technology Review
375:Ecology and Evolution
113:Circulating tumor DNA
51:conservation genetics
985:on 8 September 2018
759:2008PNAS..10519920L
753:(50): 19920–19925.
710:10.1038/nature11251
702:2012Natur.487..320F
645:2020ConGR..12..631S
496:2018EvApp..11.1094C
441:2016ConGR...8..499O
387:2020EcoEv..10.4583K
332:2019ConGR..11..465P
279:2010MolEc..19.5332P
77:cell-free fetal DNA
881:Molecular Oncology
595:10.7717/peerj.1106
541:Rethinking Ecology
85:monogenic diseases
696:(7407): 320–324.
504:10.1111/eva.12600
395:10.1002/ece3.6157
381:(11): 4583–4594.
273:(24): 5332–5344.
267:Molecular Ecology
206:10.2981/wlb.00607
173:978-0-19-514889-3
1101:
1078:
1077:
1067:
1035:
1029:
1028:
1000:
994:
993:
991:
990:
981:. Archived from
970:
964:
963:
953:
936:(224): 224ra24.
921:
915:
914:
904:
868:
862:
861:
829:
823:
822:
820:
819:
805:
799:
798:
788:
770:
738:
732:
731:
721:
681:
675:
674:
656:
624:
618:
617:
607:
597:
573:
567:
566:
556:
532:
526:
525:
515:
490:(7): 1094–1119.
475:
469:
468:
423:
417:
416:
406:
366:
360:
359:
315:
309:
308:
298:
258:
252:
251:
243:
237:
236:
218:
208:
193:Wildlife Biology
184:
178:
177:
155:
103:
63:Fetal Genotyping
1109:
1108:
1104:
1103:
1102:
1100:
1099:
1098:
1084:
1083:
1082:
1081:
1037:
1036:
1032:
1002:
1001:
997:
988:
986:
972:
971:
967:
923:
922:
918:
870:
869:
865:
831:
830:
826:
817:
815:
807:
806:
802:
740:
739:
735:
683:
682:
678:
626:
625:
621:
575:
574:
570:
534:
533:
529:
477:
476:
472:
425:
424:
420:
368:
367:
363:
317:
316:
312:
260:
259:
255:
245:
244:
240:
186:
185:
181:
174:
157:
156:
152:
147:
130:
109:
107:Tumor detection
65:
60:
43:
12:
11:
5:
1107:
1105:
1097:
1096:
1086:
1085:
1080:
1079:
1050:(4): 224–231.
1030:
1011:(3): 451–454.
995:
965:
916:
887:(3): 475–480.
875:(March 2016).
863:
824:
800:
733:
676:
639:(4): 631–641.
619:
568:
527:
470:
435:(4): 499–509.
418:
361:
326:(4): 465–471.
310:
253:
238:
179:
172:
149:
148:
146:
143:
129:
126:
108:
105:
64:
61:
59:
56:
47:3Rs principles
42:
39:
13:
10:
9:
6:
4:
3:
2:
1106:
1095:
1092:
1091:
1089:
1075:
1071:
1066:
1061:
1057:
1053:
1049:
1045:
1041:
1034:
1031:
1026:
1022:
1018:
1014:
1010:
1006:
999:
996:
984:
980:
976:
973:Standaert M.
969:
966:
961:
957:
952:
947:
943:
939:
935:
931:
927:
920:
917:
912:
908:
903:
898:
894:
890:
886:
882:
878:
874:
871:Siravegna G,
867:
864:
859:
855:
851:
847:
843:
839:
835:
828:
825:
814:
810:
804:
801:
796:
792:
787:
782:
778:
774:
769:
764:
760:
756:
752:
748:
744:
737:
734:
729:
725:
720:
715:
711:
707:
703:
699:
695:
691:
687:
680:
677:
672:
668:
664:
660:
655:
650:
646:
642:
638:
634:
630:
623:
620:
615:
611:
606:
601:
596:
591:
587:
583:
579:
572:
569:
564:
560:
555:
550:
546:
542:
538:
531:
528:
523:
519:
514:
509:
505:
501:
497:
493:
489:
485:
481:
474:
471:
466:
462:
458:
454:
450:
446:
442:
438:
434:
430:
422:
419:
414:
410:
405:
400:
396:
392:
388:
384:
380:
376:
372:
365:
362:
357:
353:
349:
345:
341:
337:
333:
329:
325:
321:
314:
311:
306:
302:
297:
292:
288:
284:
280:
276:
272:
268:
264:
257:
254:
249:
242:
239:
234:
230:
226:
222:
217:
212:
207:
202:
198:
194:
190:
183:
180:
175:
169:
165:
161:
154:
151:
144:
142:
140:
136:
127:
125:
123:
122:liquid biopsy
118:
114:
106:
104:
102:
97:
95:
94:Down Syndrome
91:
86:
82:
78:
74:
73:amniocentesis
70:
62:
57:
55:
52:
48:
40:
38:
35:
31:
27:
23:
18:
1047:
1043:
1033:
1008:
1004:
998:
987:. Retrieved
983:the original
978:
968:
933:
929:
919:
884:
880:
866:
841:
837:
827:
816:. Retrieved
812:
803:
750:
746:
736:
693:
689:
679:
636:
632:
622:
585:
581:
571:
544:
540:
530:
487:
483:
473:
432:
428:
421:
378:
374:
364:
323:
319:
313:
270:
266:
256:
241:
216:10453/143370
196:
192:
182:
163:
153:
131:
110:
98:
66:
44:
41:Conservation
16:
15:
844:(23): 114.
547:: 119–132.
30:noninvasive
989:2017-04-10
873:Bardelli A
818:2017-04-10
145:References
117:metastasis
81:aneuploidy
858:0006-4971
671:218624561
663:1877-7260
588:: e1106.
563:2534-9260
465:255785768
457:1877-7260
356:255792923
348:1877-7260
233:216291100
225:0909-6396
90:karyotype
1094:Genetics
1088:Category
1074:24294115
1025:14767578
960:24553385
911:26774880
795:19060211
777:25465749
728:22763444
614:26244114
522:30026800
413:32551045
305:21054605
135:purified
58:Medicine
34:allozyme
1065:3792701
951:4017867
902:5528968
786:2596743
755:Bibcode
719:3561905
698:Bibcode
641:Bibcode
605:4517967
513:6050181
492:Bibcode
437:Bibcode
404:7297754
383:Bibcode
328:Bibcode
296:2998560
275:Bibcode
128:Methods
1072:
1062:
1023:
958:
948:
909:
899:
856:
793:
783:
775:
726:
716:
690:Nature
669:
661:
612:
602:
561:
520:
510:
463:
455:
411:
401:
354:
346:
303:
293:
231:
223:
170:
139:primer
838:Blood
773:JSTOR
667:S2CID
582:PeerJ
461:S2CID
352:S2CID
229:S2CID
199:(1).
1070:PMID
1021:PMID
956:PMID
907:PMID
854:ISSN
791:PMID
724:PMID
659:ISSN
610:PMID
559:ISSN
518:PMID
453:ISSN
409:PMID
344:ISSN
301:PMID
221:ISSN
197:2020
168:ISBN
1060:PMC
1052:doi
1013:doi
946:PMC
938:doi
897:PMC
889:doi
846:doi
842:126
781:PMC
763:doi
751:105
714:PMC
706:doi
694:487
649:doi
600:PMC
590:doi
549:doi
508:PMC
500:doi
445:doi
399:PMC
391:doi
336:doi
291:PMC
283:doi
211:hdl
201:doi
26:DNA
22:PCR
1090::
1068:.
1058:.
1048:24
1046:.
1042:.
1019:.
1009:13
1007:.
977:.
954:.
944:.
932:.
928:.
905:.
895:.
885:10
879:.
852:.
840:.
836:.
811:.
789:.
779:.
771:.
761:.
749:.
745:.
722:.
712:.
704:.
692:.
688:.
665:.
657:.
647:.
637:12
635:.
631:.
608:.
598:.
584:.
580:.
557:.
543:.
539:.
516:.
506:.
498:.
488:11
486:.
482:.
459:.
451:.
443:.
431:.
407:.
397:.
389:.
379:10
377:.
373:.
350:.
342:.
334:.
324:11
322:.
299:.
289:.
281:.
271:19
269:.
265:.
227:.
219:.
209:.
195:.
191:.
1076:.
1054::
1027:.
1015::
992:.
962:.
940::
934:6
913:.
891::
860:.
848::
821:.
797:.
765::
757::
730:.
708::
700::
673:.
651::
643::
616:.
592::
586:3
565:.
551::
545:4
524:.
502::
494::
467:.
447::
439::
433:8
415:.
393::
385::
358:.
338::
330::
307:.
285::
277::
250:.
235:.
213::
203::
176:.
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.