107:
MuvB factors also revealed association of BMYB. Subsequent immunoprecipitation with BMYB yielded all the MuvB core proteins, but not other members of the DREAM complex – p130, p107, E2F4/5 and DP. This indicated that MuvB associated with BMYB to form the BMYB-MuvB complex or with p130/p107, E2F4/5 and DP to form the DREAM complex. The DREAM complex was found prevalent in quiescent or starved cells, and the BMYB-MuvB complex was found in actively dividing cells, hinting at separate functionalities of these two complexes.
266:
degraded and E2F4 exported from the nucleus. Once the repressive E2Fs are vacated, activating E2Fs bind to the promoter to up-regulate G1/S genes that promote DNA synthesis and transition of the cell cycle. BMYB is also up-regulated during this time, which then binds to genes that peak at G2/M phase. Binding of BMYB to late cell cycle genes is dependent on its association with the MuvB core to form the BMYB-MuvB complex, which is then able to up-regulate genes in the G2/M phase.
197:
253:
230:
via E2F proteins binding to sequences known as cell cycle-dependent element sites (CDEs). Some cell cycle dependent genes have been found where both CHRs and CDEs are in proximity to one another. Because p130-E2F4 can form stable associations with the MuvB complex, the proximity of CHRs to CDEs suggests that affinity of binding of the DREAM complex to target genes is cooperatively improved by association with both the binding sites.
229:
Docking of the DREAM complex to promoters is achieved by binding of LIN-54 to regions known as cell cycle genes homology region (CHR). These are specific sequence of nucleotides that are commonly found in the promoters of genes expressed during late S phase or G2/M phase. Docking can also be achieved
281:
Other components have been shown to be phosphorylated for DREAM complex assembly to occur. Of these, LIN52 phosphorylation on its S28 residue is the most well-understood. Substitution of this serine to alanine led to reduced binding of the MuvB core to p130 and impaired the ability of cells to enter
106:
The mammalian DREAM complex was identified following immunoprecipitation of p130 with mass-spectrometry analysis. The results showed that p130 was associated with E2F4, E2F5, the dimerization partner DP, and LIN9, LIN54, LIN37, LIN52, and RBBP4 that make up the MuvB complex. Immunoprecipitation of
265:
Like its counterpart, RB-E2F, the DREAM complex is also affected by similar growth stimuli and subsequent cyclin-CDK activity. Increasing cyclin D-CDK4 and cyclin E-CDK2 activity dissociates the DREAM complex from the promoter by phosphorylation of p130. Hyper-phosphorylated p130 is subsequently
221:
phase. Abrogation of the DREAM complex on the other hand, led to increased expression of E2F regulated genes normally repressed in the G0 phase. Contrary to mammalian cells, the fly dREAM complex was found at almost one-third of all promoters, which may reflect a broader role for dREAM in gene
83:
ovarian follicle cells identified a protein complex that bound to repeatedly amplifying chorion genes. The complex included genes that had close homology with the MuvB genes such as Mip130, Mip120 and Mip40. These Mip genes were identified as homologues of the MuvB genes LIN9, LIN54, and LIN37
780:
Kobayashi, K; Suzuki, T; Iwata, E; Nakamichi, N; Suzuki, T; Chen, P; Ohtani, M; Ishida, T; Hosoya, H; Müller, S; Leviczky, T; Pettkó-Szandtner, A; Darula, Z; Iwamoto, A; Nomoto, M; Tada, Y; Higashiyama, T; Demura, T; Doonan, JH; Hauser, MT; Sugimoto, K; Umeda, M; Magyar, Z; Bögre, L; Ito, M
830:
Lang, Lucas; Pettkó-Szandtner, Aladár; Tunçay Elbaşı, Hasibe; Takatsuka, Hirotomo; Nomoto, Yuji; Zaki, Ahmad; Dorokhov, Stefan; De Jaeger, Geert; Eeckhout, Dominique; Ito, Masaki; Magyar, Zoltán; Bögre, László; Heese, Maren; Schnittger, Arp (December 2021).
237:. Therefore, unlike RB-E2F, the DREAM complex is unlikely to directly recruit chromatin modifiers to repress gene expression, although some associations have been suggested. DREAM complex may instead down-regulate gene expression by affecting
286:. The loss of this kinase leads to decreased phosphorylation of the S28 residue and association of p130 with MuvB. DYRK1A was also found to degrade cyclin D1, which would increase p21 levels – both of which contribute to cell cycle exit.
1946:
Boichuk, S.; Parry, J. A.; Makielski, K. R.; Litovchick, L.; Baron, J. L.; Zewe, J. P.; Wozniak, A.; Mehalek, K. R.; Korzeniewski, N.; Seneviratne, D. S.; Schoffski, P.; Debiec-Rychter, M.; DeCaprio, J. A.; Duensing, A. (20 June 2013).
84:
respectively. Further studies in the fly embryo nuclear extracts confirmed the coexistence of these proteins with others such as the RB homologues Rbf1 and Rbf2, and others like E2f and Dp. The protein complex was thus termed as the
1276:
Stiegler, P; De Luca, A; Bagella, L; Giordano, A (1998-11-15). "The COOH-terminal region of pRb2/p130 binds to histone deacetylase 1 (HDAC1), enhancing transcriptional repression of the E2F-dependent cyclin A promoter".
245:. In worms for example, loss of a MuvB complex protein, LIN35, leads to loss of repressive histone associations and high expression of cell cycle dependent genes. However, direct evidence for the link between repressive
282:
quiescence. This indicates that LIN52 S28 phosphorylation is required for proper association and function of the DREAM complex via binding with p130. One known regulator of phosphorylation of the S28 residue is the
1125:
Forristal, C; Henley, SA; MacDonald, JI; Bush, JR; Ort, C; Passos, DT; Talluri, S; Ishak, CA; Thwaites, MJ; Norley, CJ; Litovchick, L; DeCaprio, JA; DiMattia, G; Holdsworth, DW; Beier, F; Dick, FA (June 2014).
138:). Entry into the cell cycle dissociates p130 from the complex and leads to subsequent recruitment of activating E2F proteins. This allows for the expression of E2F regulated late G1 and S phase genes. BMYB (
72:. When mutated, these genes produced worms with multiple vulva-like organs, hence the name ‘Muv’. Three classes of Muv genes were classified, with class B genes encoding homologues of mammalian RB,
436:
Beitel, GJ; Lambie, EJ; Horvitz, HR (2000-08-22). "The C. elegans gene lin-9, which acts in an Rb-related pathway, is required for gonadal sheath cell development and encodes a novel protein".
24:-dependent gene expression. The complex is evolutionarily conserved, although some of its components vary from species to species. In humans, the key proteins in the complex are
96:, there was also found a testis-specific paralog of the Myb-MuvB/DREAM complex known as tMAC (testis-specific meiotic arrest complex), which is involved in meiotic arrest.
92:
of the components of dREAM complex led to higher expression of E2f regulated genes that are typically silenced, implicating dREAM’s role in gene down-regulation. Later in
1229:"Deletion of the p107/p130-binding domain of Mip130/LIN-9 bypasses the requirement for CDK4 activity for the dissociation of Mip130/LIN-9 from p107/p130-E2F4 complex"
278:. Inhibition of PP2a activity reduced promoter binding of some of the proteins of the DREAM complex in the subsequent G1 phase and de-repression of gene expression.
103:
extract containing DP, RB, and MuvB, and was named as DRM. This complex included mammalian homologues of RB and DP, and other members of the MuvB complex.
1758:
Naetar, N; Soundarapandian, V; Litovchick, L; Goguen, KL; Sablina, AA; Bowman-Colin, C; Sicinski, P; Hahn, WC; DeCaprio, JA; Livingston, DM (2014-06-19).
986:"Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells"
1082:
Müller, GA; Engeland, K (February 2010). "The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription".
1991:
Rashid, NN; Yusof, R; Watson, RJ (November 2014). "A B-myb--DREAM complex is not critical to regulate the G2/M genes in HPV-transformed cell lines".
1035:"The dREAM/Myb–MuvB complex and Grim are key regulators of the programmed death of neural precursor cells at the Drosophila posterior wing margin"
675:"Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex"
739:
Litovchick, L; Sadasivam, S; Florens, L; Zhu, X; Swanson, SK; Velmurugan, S; Chen, R; Washburn, MP; Liu, XS; DeCaprio, JA (2007-05-25).
577:"Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes"
575:
Korenjak, M; Taylor-Harding, B; Binné, UK; Satterlee, JS; Stevaux, O; Aasland, R; White-Cooper, H; Dyson, N; Brehm, A (2004-10-15).
741:"Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence"
1390:"The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2)"
984:
Georlette, D; Ahn, S; MacAlpine, DM; Cheung, E; Lewis, PW; Beall, EL; Bell, SP; Speed, T; Manak, JR; Botchan, MR (2007-11-15).
1537:"Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression"
1584:
Lam, EW; Robinson, C; Watson, RJ (September 1992). "Characterization and cell cycle-regulated expression of mouse B-myb".
210:
29:
25:
1949:"The DREAM Complex Mediates GIST Cell Quiescence and Is a Novel Therapeutic Target to Enhance Imatinib-Induced Apoptosis"
301:
Due to its regulatory role in the cell cycle, targeting the DREAM complex might enhance anticancer treatments such as
524:; Botchan, MR (2002-12-19). "Role for a Drosophila Myb-containing protein complex in site-specific DNA replication".
1809:"Dosage of Dyrk1a shifts cells within a p21-cyclin D1 signaling map to control the decision to enter the cell cycle"
1339:
Latorre, I; Chesney, MA; Garrigues, JM; Stempor, P; Appert, A; Francesconi, M; Strome, S; Ahringer, J (2015-03-01).
274:
Near the end of mitosis, p130 and p107 are dephosphorylated from their hyperphosphorylated state by the phosphatase
2021:
218:
127:
1711:"PP2A: more than a reset switch to activate pRB proteins during the cell cycle and in response to signaling cues"
473:"New genes that interact with lin-35 Rb to negatively regulate the let-60 ras pathway in Caenorhabditis elegans"
2031:
233:
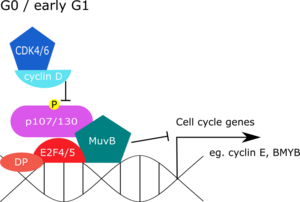
When DREAM is docked onto the promoter, p130 is bound to LIN52, and this association inhibits LIN52 binding to
193:. Near the end of the cell cycle, the DREAM complex is re-assembled by DYRK1A to repress G1/S and G2/M genes.
1856:
Wolter, P; Schmitt, K; Fackler, M; Kremling, H; Probst, L; Hauser, S; Gruss, OJ; Gaubatz, S (15 May 2012).
241:
positioning. Compacted DNA at transcription start sites inhibit gene expression by blocking the docking of
33:
1760:"PP2A-mediated regulation of Ras signaling in G2 is essential for stable quiescence and normal G1 length"
76:, and DP1, and others such as LIN-54, LIN-37, LIN-7 and LIN-52, whose functions were not yet understood.
314:
275:
223:
1858:"GAS2L3, a target gene of the DREAM complex, is required for proper cytokinesis and genomic stability"
686:
533:
234:
68:
Genes encoding the MuvB complex were originally identified from loss-of-function mutation studies in
521:
209:
During quiescence, the DREAM complex represses G1/S and G2/M gene expression. In mammalian systems,
2026:
213:
studies have revealed that DREAM components are found together at promoters of genes that peak in
1899:"The DREAM complex in antitumor activity of imatinib mesylate in gastrointestinal stromal tumors"
1691:
1662:"PP1 and PP2A phosphatases--cooperating partners in modulating retinoblastoma protein activation"
1642:
1468:
1107:
606:
557:
418:
626:"Discovery of tMAC: a Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B"
196:
1613:"Mip/LIN-9 regulates the expression of B-Myb and the induction of cyclin A, cyclin B, and CDK1"
1033:
Rovani, Margritte K.; Brachmann, Carrie Baker; Ramsay, Gary; Katzen, Alisa L. (December 2012).
624:
Beall, E. L.; Lewis, P. W.; Bell, M.; Rocha, M.; Jones, D. L.; Botchan, M. R. (15 April 2007).
2000:
1970:
1928:
1879:
1838:
1789:
1740:
1683:
1634:
1593:
1566:
1517:
1460:
1439:"SKP2 associates with p130 and accelerates p130 ubiquitylation and degradation in human cells"
1419:
1370:
1321:
1286:
1258:
1209:
1157:
1099:
1064:
1015:
966:
914:
862:
812:
762:
714:
655:
598:
549:
502:
453:
410:
369:
1960:
1918:
1910:
1869:
1828:
1820:
1779:
1771:
1730:
1722:
1673:
1624:
1556:
1548:
1507:
1499:
1450:
1409:
1401:
1360:
1352:
1313:
1248:
1240:
1199:
1191:
1147:
1139:
1091:
1054:
1046:
1005:
997:
956:
948:
904:
896:
852:
844:
802:
794:
752:
704:
694:
645:
637:
588:
541:
492:
484:
445:
400:
359:
351:
89:
885:"The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression"
252:
1178:
Guiley, KZ; Liban, TJ; Felthousen, JG; Ramanan, P; Litovchick, L; Rubin, SM (2015-05-01).
214:
123:
937:"DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly"
690:
537:
1923:
1898:
1833:
1808:
1784:
1759:
1735:
1710:
1365:
1340:
1253:
1228:
1204:
1179:
1152:
1127:
1059:
1034:
1010:
985:
961:
936:
909:
884:
857:
832:
807:
782:
709:
674:
650:
625:
497:
472:
364:
339:
242:
1561:
1536:
1414:
1389:
449:
2015:
1678:
1661:
1512:
1487:
1095:
422:
1695:
1646:
1503:
1472:
610:
1437:
Bhattacharya, S; Garriga, J; Calbó, J; Yong, T; Haines, DS; Graña, X (2003-04-24).
1111:
561:
1965:
1948:
1304:
Bai, L; Morozov, AV (November 2010). "Gene regulation by nucleosome positioning".
935:
Litovchick, L; Florens, LA; Swanson, SK; Washburn, MP; DeCaprio, JA (2011-04-15).
405:
388:
1914:
1824:
1775:
1726:
757:
740:
488:
1244:
1050:
290:
186:
679:
Proceedings of the
National Academy of Sciences of the United States of America
593:
576:
340:"The DREAM complex: master coordinator of cell cycle-dependent gene expression"
1317:
238:
21:
833:"The DREAM complex represses growth in response to DNA damage in Arabidopsis"
798:
699:
2004:
1974:
1932:
1883:
1842:
1793:
1744:
1687:
1638:
1629:
1612:
1611:
Pilkinton, M; Sandoval, R; Song, J; Ness, SA; Colamonici, OR (2007-01-05).
1570:
1521:
1464:
1455:
1438:
1423:
1374:
1356:
1325:
1262:
1213:
1195:
1161:
1128:"Loss of the mammalian DREAM complex deregulates chondrocyte proliferation"
1103:
1068:
1019:
970:
918:
900:
866:
848:
816:
783:"Transcriptional repression by MYB3R proteins regulates plant organ growth"
766:
718:
659:
602:
553:
506:
457:
414:
373:
114:
that include E2F and MYB orthologs combined with LIN9 and LIN54 orthologs.
88:
RBF, E2f2 and Mip (dREAM) complex. Disruption of the dREAM complex through
18:
dimerization partner, RB-like, E2F and multi-vulval class B (DREAM) complex
1597:
1290:
1552:
1143:
302:
151:
131:
1405:
1001:
952:
641:
545:
1874:
1857:
246:
158:
147:
52:
and DP3, dimerization partners of E2F; and MuvB, which is a complex of
283:
1486:
Gaubatz, S; Lees, JA; Lindeman, GJ; Livingston, DM (February 2001).
355:
146:
is also able to be expressed at this time, and binds to MuvB during
389:"Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes"
1341:"The DREAM complex promotes gene body H2A.Z for target repression"
190:
178:
170:
166:
139:
99:
A protein complex similar to dREAM was subsequently identified in
57:
49:
45:
1180:"Structural mechanisms of DREAM complex assembly and regulation"
182:
162:
53:
41:
37:
1488:"E2F4 is exported from the nucleus in a CRM1-dependent manner"
73:
471:
Thomas, JH; Ceol, CJ; Schwartz, HT; Horvitz, HR (May 2003).
338:
Sadasivam, Subhashini; DeCaprio, James A. (11 July 2013).
883:
Sadasivam, S.; Duan, S.; DeCaprio, J. A. (5 March 2012).
673:
Harrison, MM; Ceol, CJ; Lu, X; Horvitz, HR (2006-11-07).
1227:
Sandoval, R; Pilkinton, M; Colamonici, OR (2009-10-15).
20:
is a protein complex responsible for the regulation of
393:
Critical
Reviews in Biochemistry and Molecular Biology
1535:
Takahashi, Y; Rayman, JB; Dynlacht, BD (2000-04-01).
122:
The main function of the DREAM complex is to repress
110:
MuvB-like complexes were also recently discovered in
36:(p105) and bind repressive E2F transcription factors
1807:
Chen, JY; Lin, JR; Tsai, FC; Meyer, T (2013-10-10).
142:), which is repressed by the DREAM complex during G
1897:DeCaprio, James A.; Duensing, Anette (July 2014).
930:
928:
878:
876:
734:
732:
730:
728:
189:), while FOXM1 is degraded during mitosis by the
249:and the DREAM complex remains to be elucidated.
1173:
1171:
1388:Tedesco, D; Lukas, J; Reed, SI (2002-11-15).
289:The DREAM complex was also shown to regulate
8:
333:
331:
329:
181:). During late S phase BMYB is degraded via
256:Mammalian DREAM complex disassembly in G1/S
1660:Kolupaeva, V; Janssens, V (January 2013).
200:Mammalian DREAM complex in G0 and early G1
1964:
1922:
1873:
1832:
1783:
1734:
1677:
1628:
1560:
1511:
1454:
1413:
1364:
1252:
1203:
1151:
1058:
1009:
960:
908:
856:
806:
756:
708:
698:
649:
592:
496:
404:
363:
177:to further promote gene expression (e.g.
520:Beall, EL; Manak, JR; Zhou, S; Bell, M;
387:Fischer, M; Müller, GA (December 2017).
251:
195:
325:
32:(p130), both of which are homologs of
7:
211:chromatin-immunoprecipitation (ChIP)
1617:The Journal of Biological Chemistry
14:
150:to promote the expression of key
1679:10.1111/j.1742-4658.2012.08511.x
1096:10.1111/j.1742-4658.2009.07508.x
1709:Kurimchak, A; Graña, X (2015).
1504:10.1128/MCB.21.4.1384-1392.2001
1492:Molecular and Cellular Biology
1132:Molecular and Cellular Biology
1:
1966:10.1158/0008-5472.CAN-13-0579
450:10.1016/s0378-1119(00)00296-1
406:10.1080/10409238.2017.1360836
1915:10.1097/CCO.0000000000000090
1825:10.1016/j.molcel.2013.09.009
1776:10.1016/j.molcel.2014.04.023
1727:10.4161/15384101.2014.985069
758:10.1016/j.molcel.2007.04.015
1903:Current Opinion in Oncology
1245:10.1016/j.yexcr.2009.07.014
1051:10.1016/j.ydbio.2012.08.022
235:chromatin modifier proteins
226:of neural precursor cells.
2048:
1233:Experimental Cell Research
594:10.1016/j.cell.2004.09.034
489:10.1093/genetics/164.1.135
1318:10.1016/j.tig.2010.08.003
1862:Journal of Cell Science
1541:Genes & Development
1394:Genes & Development
1345:Genes & Development
1184:Genes & Development
990:Genes & Development
941:Genes & Development
889:Genes & Development
799:10.15252/embj.201490899
700:10.1073/pnas.0608461103
630:Genes & Development
130:gene expression during
94:Drosophila melanogaster
81:Drosophila melanogaster
1630:10.1074/jbc.M609924200
1456:10.1038/sj.onc.1206339
1357:10.1101/gad.255810.114
1196:10.1101/gad.257568.114
901:10.1101/gad.181933.111
849:10.26508/lsa.202101141
257:
201:
173:is then recruited in G
1039:Developmental Biology
837:Life Science Alliance
344:Nature Reviews Cancer
315:Pocket protein family
255:
224:programmed cell death
199:
1553:10.1101/gad.14.7.804
1144:10.1128/MCB.01523-13
222:regulation, such as
161:phase genes such as
1993:Anticancer Research
1868:(Pt 10): 2393–406.
1406:10.1101/gad.1011202
1002:10.1101/gad.1600107
953:10.1101/gad.2034211
691:2006PNAS..10316782H
642:10.1101/gad.1516607
546:10.1038/nature01228
538:2002Natur.420..833B
1875:10.1242/jcs.097253
1306:Trends in Genetics
843:(12): e202101141.
258:
202:
2022:Protein complexes
1959:(16): 5120–5129.
793:(15): 1992–2007.
2039:
2008:
1979:
1978:
1968:
1943:
1937:
1936:
1926:
1894:
1888:
1887:
1877:
1853:
1847:
1846:
1836:
1804:
1798:
1797:
1787:
1755:
1749:
1748:
1738:
1706:
1700:
1699:
1681:
1666:The FEBS Journal
1657:
1651:
1650:
1632:
1608:
1602:
1601:
1581:
1575:
1574:
1564:
1532:
1526:
1525:
1515:
1483:
1477:
1476:
1458:
1434:
1428:
1427:
1417:
1385:
1379:
1378:
1368:
1336:
1330:
1329:
1301:
1295:
1294:
1273:
1267:
1266:
1256:
1224:
1218:
1217:
1207:
1175:
1166:
1165:
1155:
1122:
1116:
1115:
1084:The FEBS Journal
1079:
1073:
1072:
1062:
1030:
1024:
1023:
1013:
981:
975:
974:
964:
932:
923:
922:
912:
880:
871:
870:
860:
827:
821:
820:
810:
787:The EMBO Journal
777:
771:
770:
760:
736:
723:
722:
712:
702:
670:
664:
663:
653:
621:
615:
614:
596:
572:
566:
565:
517:
511:
510:
500:
468:
462:
461:
433:
427:
426:
408:
384:
378:
377:
367:
335:
293:through GAS2L3.
2047:
2046:
2042:
2041:
2040:
2038:
2037:
2036:
2032:Gene expression
2012:
2011:
1999:(11): 6557–63.
1990:
1987:
1985:Further reading
1982:
1953:Cancer Research
1945:
1944:
1940:
1896:
1895:
1891:
1855:
1854:
1850:
1806:
1805:
1801:
1757:
1756:
1752:
1708:
1707:
1703:
1659:
1658:
1654:
1610:
1609:
1605:
1583:
1582:
1578:
1534:
1533:
1529:
1485:
1484:
1480:
1449:(16): 2443–51.
1436:
1435:
1431:
1400:(22): 2946–57.
1387:
1386:
1382:
1338:
1337:
1333:
1303:
1302:
1298:
1285:(22): 5049–52.
1279:Cancer Research
1275:
1274:
1270:
1239:(17): 2914–20.
1226:
1225:
1221:
1177:
1176:
1169:
1138:(12): 2221–34.
1124:
1123:
1119:
1081:
1080:
1076:
1032:
1031:
1027:
996:(22): 2880–96.
983:
982:
978:
934:
933:
926:
882:
881:
874:
829:
828:
824:
779:
778:
774:
738:
737:
726:
685:(45): 16782–7.
672:
671:
667:
623:
622:
618:
574:
573:
569:
532:(6917): 833–7.
519:
518:
514:
470:
469:
465:
444:(1–2): 253–63.
435:
434:
430:
386:
385:
381:
356:10.1038/nrc3556
337:
336:
327:
323:
311:
299:
272:
263:
207:
176:
155:
145:
137:
120:
66:
12:
11:
5:
2045:
2043:
2035:
2034:
2029:
2024:
2014:
2013:
2010:
2009:
1986:
1983:
1981:
1980:
1938:
1909:(4): 415–421.
1889:
1848:
1813:Molecular Cell
1799:
1764:Molecular Cell
1750:
1701:
1652:
1603:
1592:(9): 1885–90.
1576:
1527:
1498:(4): 1384–92.
1478:
1429:
1380:
1351:(5): 495–500.
1331:
1312:(11): 476–83.
1296:
1268:
1219:
1167:
1117:
1074:
1025:
976:
924:
895:(5): 474–489.
872:
822:
781:(2015-08-04).
772:
745:Molecular Cell
724:
665:
636:(8): 904–919.
616:
567:
512:
463:
428:
399:(6): 638–662.
379:
350:(8): 585–595.
324:
322:
319:
318:
317:
310:
307:
298:
297:Cancer therapy
295:
271:
268:
262:
259:
243:RNA polymerase
206:
203:
174:
153:
143:
135:
119:
116:
90:RNAi knockdown
65:
62:
56:/37/52/54 and
13:
10:
9:
6:
4:
3:
2:
2044:
2033:
2030:
2028:
2025:
2023:
2020:
2019:
2017:
2006:
2002:
1998:
1994:
1989:
1988:
1984:
1976:
1972:
1967:
1962:
1958:
1954:
1950:
1942:
1939:
1934:
1930:
1925:
1920:
1916:
1912:
1908:
1904:
1900:
1893:
1890:
1885:
1881:
1876:
1871:
1867:
1863:
1859:
1852:
1849:
1844:
1840:
1835:
1830:
1826:
1822:
1819:(1): 87–100.
1818:
1814:
1810:
1803:
1800:
1795:
1791:
1786:
1781:
1777:
1773:
1770:(6): 932–45.
1769:
1765:
1761:
1754:
1751:
1746:
1742:
1737:
1732:
1728:
1724:
1720:
1716:
1712:
1705:
1702:
1697:
1693:
1689:
1685:
1680:
1675:
1672:(2): 627–43.
1671:
1667:
1663:
1656:
1653:
1648:
1644:
1640:
1636:
1631:
1626:
1623:(1): 168–75.
1622:
1618:
1614:
1607:
1604:
1599:
1595:
1591:
1587:
1580:
1577:
1572:
1568:
1563:
1558:
1554:
1550:
1547:(7): 804–16.
1546:
1542:
1538:
1531:
1528:
1523:
1519:
1514:
1509:
1505:
1501:
1497:
1493:
1489:
1482:
1479:
1474:
1470:
1466:
1462:
1457:
1452:
1448:
1444:
1440:
1433:
1430:
1425:
1421:
1416:
1411:
1407:
1403:
1399:
1395:
1391:
1384:
1381:
1376:
1372:
1367:
1362:
1358:
1354:
1350:
1346:
1342:
1335:
1332:
1327:
1323:
1319:
1315:
1311:
1307:
1300:
1297:
1292:
1288:
1284:
1280:
1272:
1269:
1264:
1260:
1255:
1250:
1246:
1242:
1238:
1234:
1230:
1223:
1220:
1215:
1211:
1206:
1201:
1197:
1193:
1190:(9): 961–74.
1189:
1185:
1181:
1174:
1172:
1168:
1163:
1159:
1154:
1149:
1145:
1141:
1137:
1133:
1129:
1121:
1118:
1113:
1109:
1105:
1101:
1097:
1093:
1090:(4): 877–93.
1089:
1085:
1078:
1075:
1070:
1066:
1061:
1056:
1052:
1048:
1045:(1): 88–102.
1044:
1040:
1036:
1029:
1026:
1021:
1017:
1012:
1007:
1003:
999:
995:
991:
987:
980:
977:
972:
968:
963:
958:
954:
950:
947:(8): 801–13.
946:
942:
938:
931:
929:
925:
920:
916:
911:
906:
902:
898:
894:
890:
886:
879:
877:
873:
868:
864:
859:
854:
850:
846:
842:
838:
834:
826:
823:
818:
814:
809:
804:
800:
796:
792:
788:
784:
776:
773:
768:
764:
759:
754:
751:(4): 539–51.
750:
746:
742:
735:
733:
731:
729:
725:
720:
716:
711:
706:
701:
696:
692:
688:
684:
680:
676:
669:
666:
661:
657:
652:
647:
643:
639:
635:
631:
627:
620:
617:
612:
608:
604:
600:
595:
590:
587:(2): 181–93.
586:
582:
578:
571:
568:
563:
559:
555:
551:
547:
543:
539:
535:
531:
527:
523:
516:
513:
508:
504:
499:
494:
490:
486:
483:(1): 135–51.
482:
478:
474:
467:
464:
459:
455:
451:
447:
443:
439:
432:
429:
424:
420:
416:
412:
407:
402:
398:
394:
390:
383:
380:
375:
371:
366:
361:
357:
353:
349:
345:
341:
334:
332:
330:
326:
320:
316:
313:
312:
308:
306:
304:
296:
294:
292:
287:
285:
279:
277:
269:
267:
260:
254:
250:
248:
244:
240:
236:
231:
227:
225:
220:
216:
212:
204:
198:
194:
192:
188:
184:
180:
172:
168:
164:
160:
156:
149:
141:
133:
129:
125:
117:
115:
113:
108:
104:
102:
97:
95:
91:
87:
82:
77:
75:
71:
63:
61:
59:
55:
51:
47:
43:
39:
35:
31:
27:
23:
19:
1996:
1992:
1956:
1952:
1941:
1906:
1902:
1892:
1865:
1861:
1851:
1816:
1812:
1802:
1767:
1763:
1753:
1721:(1): 18–30.
1718:
1714:
1704:
1669:
1665:
1655:
1620:
1616:
1606:
1589:
1585:
1579:
1544:
1540:
1530:
1495:
1491:
1481:
1446:
1442:
1432:
1397:
1393:
1383:
1348:
1344:
1334:
1309:
1305:
1299:
1282:
1278:
1271:
1236:
1232:
1222:
1187:
1183:
1135:
1131:
1120:
1087:
1083:
1077:
1042:
1038:
1028:
993:
989:
979:
944:
940:
892:
888:
840:
836:
825:
790:
786:
775:
748:
744:
682:
678:
668:
633:
629:
619:
584:
580:
570:
529:
525:
515:
480:
476:
466:
441:
437:
431:
396:
392:
382:
347:
343:
300:
288:
280:
273:
270:Late mitosis
264:
232:
228:
208:
121:
112:Arabidoposis
111:
109:
105:
100:
98:
93:
85:
80:
78:
69:
67:
17:
15:
522:Lipsick, JS
291:cytokinesis
187:SCF complex
79:Studies in
28:(p107) and
2027:Cell cycle
2016:Categories
1715:Cell Cycle
321:References
239:nucleosome
132:quiescence
101:C. elegans
86:Drosophila
70:C. elegans
22:cell cycle
423:205695213
64:Discovery
2005:25368258
1975:23786773
1933:24840522
1884:22344256
1843:24119401
1794:24857551
1745:25483052
1696:46705471
1688:22299668
1647:21963932
1639:17098733
1586:Oncogene
1571:10766737
1522:11158323
1473:26125392
1465:12717421
1443:Oncogene
1424:12435635
1375:25737279
1326:20832136
1263:19619530
1214:25917549
1162:24710275
1104:20015071
1069:22960039
1020:17978103
971:21498570
919:22391450
867:34583930
817:26069325
767:17531812
719:17075059
660:17403774
611:17989678
603:15479636
554:12490953
507:12750327
477:Genetics
458:10974557
415:28799433
374:23842645
309:See also
303:imatinib
247:histones
118:Function
1924:4236229
1834:4039290
1785:4118046
1736:4612414
1598:1501895
1366:4358402
1291:9823308
1254:2757496
1205:4421984
1153:4054284
1112:8955433
1060:3621911
1011:2049191
962:3078706
910:3305985
858:8500230
808:4551348
710:1636532
687:Bibcode
651:1847709
562:4425307
534:Bibcode
498:1462563
365:3986830
148:S phase
2003:
1973:
1931:
1921:
1882:
1841:
1831:
1792:
1782:
1743:
1733:
1694:
1686:
1645:
1637:
1596:
1569:
1562:316494
1559:
1520:
1510:
1471:
1463:
1422:
1415:187481
1412:
1373:
1363:
1324:
1289:
1261:
1251:
1212:
1202:
1160:
1150:
1110:
1102:
1067:
1057:
1018:
1008:
969:
959:
917:
907:
865:
855:
815:
805:
765:
717:
707:
658:
648:
609:
601:
560:
552:
526:Nature
505:
495:
456:
421:
413:
372:
362:
284:DYRK1A
1692:S2CID
1643:S2CID
1513:99590
1469:S2CID
1108:S2CID
607:S2CID
558:S2CID
419:S2CID
191:APC/C
179:AURKA
171:FOXM1
167:CCNB1
140:MYBL2
58:RBBP4
2001:PMID
1971:PMID
1929:PMID
1880:PMID
1839:PMID
1790:PMID
1741:PMID
1684:PMID
1635:PMID
1594:PMID
1567:PMID
1518:PMID
1461:PMID
1420:PMID
1371:PMID
1322:PMID
1287:PMID
1259:PMID
1210:PMID
1158:PMID
1100:PMID
1065:PMID
1016:PMID
967:PMID
915:PMID
863:PMID
813:PMID
763:PMID
715:PMID
656:PMID
599:PMID
581:Cell
550:PMID
503:PMID
454:PMID
438:Gene
411:PMID
370:PMID
276:PP2a
261:G1/S
219:G2/M
215:G1/S
183:CUL1
165:and
163:CDK1
128:G2/M
126:and
124:G1/S
54:LIN9
42:E2F5
40:and
38:E2F4
30:RBL2
26:RBL1
16:The
1961:doi
1919:PMC
1911:doi
1870:doi
1866:125
1829:PMC
1821:doi
1780:PMC
1772:doi
1731:PMC
1723:doi
1674:doi
1670:280
1625:doi
1621:282
1557:PMC
1549:doi
1508:PMC
1500:doi
1451:doi
1410:PMC
1402:doi
1361:PMC
1353:doi
1314:doi
1249:PMC
1241:doi
1237:315
1200:PMC
1192:doi
1148:PMC
1140:doi
1092:doi
1088:277
1055:PMC
1047:doi
1043:372
1006:PMC
998:doi
957:PMC
949:doi
905:PMC
897:doi
853:PMC
845:doi
803:PMC
795:doi
753:doi
705:PMC
695:doi
683:103
646:PMC
638:doi
589:doi
585:119
542:doi
530:420
493:PMC
485:doi
481:164
446:doi
442:254
401:doi
360:PMC
352:doi
217:or
74:E2F
50:DP2
46:DP1
2018::
1997:34
1995:.
1969:.
1957:73
1955:.
1951:.
1927:.
1917:.
1907:26
1905:.
1901:.
1878:.
1864:.
1860:.
1837:.
1827:.
1817:52
1815:.
1811:.
1788:.
1778:.
1768:54
1766:.
1762:.
1739:.
1729:.
1719:14
1717:.
1713:.
1690:.
1682:.
1668:.
1664:.
1641:.
1633:.
1619:.
1615:.
1588:.
1565:.
1555:.
1545:14
1543:.
1539:.
1516:.
1506:.
1496:21
1494:.
1490:.
1467:.
1459:.
1447:22
1445:.
1441:.
1418:.
1408:.
1398:16
1396:.
1392:.
1369:.
1359:.
1349:29
1347:.
1343:.
1320:.
1310:26
1308:.
1283:58
1281:.
1257:.
1247:.
1235:.
1231:.
1208:.
1198:.
1188:29
1186:.
1182:.
1170:^
1156:.
1146:.
1136:34
1134:.
1130:.
1106:.
1098:.
1086:.
1063:.
1053:.
1041:.
1037:.
1014:.
1004:.
994:21
992:.
988:.
965:.
955:.
945:25
943:.
939:.
927:^
913:.
903:.
893:26
891:.
887:.
875:^
861:.
851:.
839:.
835:.
811:.
801:.
791:34
789:.
785:.
761:.
749:26
747:.
743:.
727:^
713:.
703:.
693:.
681:.
677:.
654:.
644:.
634:21
632:.
628:.
605:.
597:.
583:.
579:.
556:.
548:.
540:.
528:.
501:.
491:.
479:.
475:.
452:.
440:.
417:.
409:.
397:52
395:.
391:.
368:.
358:.
348:13
346:.
342:.
328:^
305:.
205:G0
169:.
134:(G
60:.
48:,
44:;
34:RB
2007:.
1977:.
1963::
1935:.
1913::
1886:.
1872::
1845:.
1823::
1796:.
1774::
1747:.
1725::
1698:.
1676::
1649:.
1627::
1600:.
1590:7
1573:.
1551::
1524:.
1502::
1475:.
1453::
1426:.
1404::
1377:.
1355::
1328:.
1316::
1293:.
1265:.
1243::
1216:.
1194::
1164:.
1142::
1114:.
1094::
1071:.
1049::
1022:.
1000::
973:.
951::
921:.
899::
869:.
847::
841:4
819:.
797::
769:.
755::
721:.
697::
689::
662:.
640::
613:.
591::
564:.
544::
536::
509:.
487::
460:.
448::
425:.
403::
376:.
354::
185:(
175:2
159:M
157:/
154:2
152:G
144:0
136:0
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.