575:
3374:
242:
43:
708:
often challenging, because only very few genes show equal levels of expression across a range of different conditions or tissues. Although cycle threshold analysis is integrated with many commercial software systems, there are more accurate and reliable methods of analysing amplification profile data that should be considered in cases where reproducibility is a concern.
878:
amplification. Using taxonomic markers (ribosomal genes) and qPCR can help determine the amount of microorganisms in a sample, and can identify different families, genera, or species based on the specificity of the marker. Using functional markers (protein-coding genes) can show gene expression within a community, which may reveal information about the environment.
151:
140:
491:, and after each cycle, the intensity of fluorescence is measured with a detector; the dye only fluoresces when bound to the dsDNA (i.e., the PCR product). This method has the advantage of only needing a pair of primers to carry out the amplification, which keeps costs down; multiple target sequences can be monitored in a tube by using different types of dyes.
325:) in several samples. Quantitative PCR can also be applied to the detection and quantification of DNA in samples to determine the presence and abundance of a particular DNA sequence in these samples. This measurement is made after each amplification cycle, and this is the reason why this method is called real time PCR (that is, immediate or simultaneous PCR).
712:
use knowledge about the polymerase amplification process to generate estimates of the original sample concentration. An extension of this approach includes an accurate model of the entire PCR reaction profile, which allows for the use of high signal-to-noise data and the ability to validate data quality prior to analysis.
373:) have been developed. Some have been developed for quantifying total gene expression, but the most common are aimed at quantifying the specific gene being studied in relation to another gene called a normalizing gene, which is selected for its almost constant level of expression. These genes are often selected from
715:
According to research of
Ruijter et al. MAK2 assumes constant amplification efficiency during the PCR reaction. However, theoretical analysis of polymerase chain reaction, from which MAK2 was derived, has revealed that amplification efficiency is not constant throughout PCR. While MAK2 quantification
578:
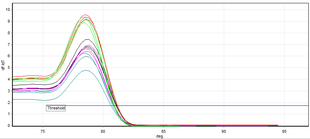
Distinct fusion curves for a number of PCR products (showing distinct colours). Amplification reactions can be seen for a specific product (pink, blue) and others with a negative result (green, orange). The fusion peak indicated with an arrow shows the peak caused by primer dimers, which is different
441:
The PCR process generally consists of a series of temperature changes that are repeated 25–50 times. These cycles normally consist of three stages: the first, at around 95 °C, allows the separation of the nucleic acid's double chain; the second, at a temperature of around 50–60 °C, allows the binding
360:
to the gene of interest. Although this technique is still used to assess gene expression, it requires relatively large amounts of RNA and provides only qualitative or semi quantitative information of mRNA levels. Estimation errors arising from variations in the quantification method can be the result
602:). The method used is usually PCR with double-stranded DNA-binding dyes as reporters and the dye used is usually SYBR Green. The DNA melting temperature is specific to the amplified fragment. The results of this technique are obtained by comparing the dissociation curves of the analysed DNA samples.
450:
during the change between the alignment stage and the denaturing stage. In addition, in four-step PCR the fluorescence is measured during short temperature phases lasting only a few seconds in each cycle, with a temperature of, for example, 80 °C, in order to reduce the signal caused by the presence
921:
or even intermediate sequences used during the process of engineering the vector. As the process of creating a transgenic plant normally leads to the insertion of more than one copy of the transgene its quantity is also commonly assessed. This is often carried out by relative quantification using a
711:
Mechanism-based qPCR quantification methods have also been suggested, and have the advantage that they do not require a standard curve for quantification. Methods such as MAK2 have been shown to have equal or better quantitative performance to standard curve methods. These mechanism-based methods
510:
reporter probes detect only the DNA containing the sequence complementary to the probe; therefore, use of the reporter probe significantly increases specificity, and enables performing the technique even in the presence of other dsDNA. Using different-coloured labels, fluorescent probes can be used
794:
As the units used to express the results of relative quantification are unimportant the results can be compared across a number of different RTqPCR. The reason for using one or more housekeeping genes is to correct non-specific variation, such as the differences in the quantity and quality of RNA
707:
and permits comparison of expression of a gene of interest among different samples. However, for such comparison, expression of the normalizing reference gene needs to be very similar across all the samples. Choosing a reference gene fulfilling this criterion is therefore of high importance, and
609:
techniques to demonstrate the results of all the samples. This is because, despite being a kinetic technique, quantitative PCR is usually evaluated at a distinct end point. The technique therefore usually provides more rapid results and/or uses fewer reactants than electrophoresis. If subsequent
877:
qPCR may also be used to amplify taxonomic or functional markers of genes in DNA taken from environmental samples. Markers are represented by genetic fragments of DNA or complementary DNA. By amplifying a certain genetic element, one can quantify the amount of the element in the sample prior to
618:
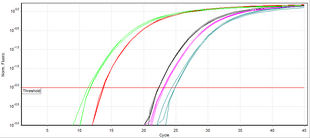
Unlike end point PCR (conventional PCR), real time PCR allows monitoring of the desired product at any point in the amplification process by measuring fluorescence (in real time frame, measurement is made of its level over a given threshold). A commonly employed method of DNA quantification by
265:): The amount of an expressed gene in a cell can be measured by the number of copies of an RNA transcript of that gene present in a sample. In order to robustly detect and quantify gene expression from small amounts of RNA, amplification of the gene transcript is necessary. The
1685:
Ruijter JM, Pfaffl MW, Zhao S, Spiess AN, Boggy G, Blom J, Rutledge RG, Sisti D, Lievens A, De Preter K, Derveaux S, Hellemans J, Vandesompele J (2012). "Evaluation of qPCR curve analysis methods for reliable biomarker discovery: bias, resolution, precision, and implications".
503:(1) In intact probes, reporter fluorescence is quenched. (2) Probes and the complementary DNA strand are hybridized and reporter fluorescence is still quenched. (3) During PCR, the probe is degraded by the Taq polymerase and the fluorescent reporter released.
886:
The agricultural industry is constantly striving to produce plant propagules or seedlings that are free of pathogens in order to prevent economic losses and safeguard health. Systems have been developed that allow detection of small amounts of the DNA of
316:
allowing the generation rate to be measured for one or more specific products. This allows the rate of generation of the amplified product to be measured at each PCR cycle. The data thus generated can be analysed by computer software to calculate
679:
difference of 1. The cycle threshold method makes several assumptions of reaction mechanism and has a reliance on data from low signal-to-noise regions of the amplification profile that can introduce substantial variance during the data analysis.
646:
precedes that of another sample by 3 cycles contained 2 = 8 times more template. However, the efficiency of amplification is often variable among primers and templates. Therefore, the efficiency of a primer-template combination is assessed in a
553:
Polymerisation of a new DNA strand is initiated from the primers, and once the polymerase reaches the probe, its 5'-3'-exonuclease degrades the probe, physically separating the fluorescent reporter from the quencher, resulting in an increase in
385:. This enables researchers to report a ratio for the expression of the genes of interest divided by the expression of the selected normalizer, thereby allowing comparison of the former without actually knowing its absolute level of expression.
500:
873:
Quantitative PCR is also used by microbiologists working in the fields of food safety, food spoilage and fermentation and for the microbial risk assessment of water quality (drinking and recreational waters) and in public health protection.
451:
of primer dimers when a non-specific dye is used. The temperatures and the timings used for each cycle depend on a wide variety of parameters, such as: the enzyme used to synthesize the DNA, the concentration of divalent ions and
2353:
Bouchez, Blieux, Dequiedt, Domaizon, Dufresne, Ferreira, Godon, Hellal, Joulian, Quaiser, Martin-Laurent, Mauffret, Monier, Peyret, Schmitt-Koplin, Sibourg, D’oiron, Bispo, Deportes, Grand, Cuny, Maron, Ranjard (September 2016).
1081:
Pfaffl, MW; Tichopad, A; Prgomet, C; Neuvians, TP (March 2004). "Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations".
1722:
814:. However, research has shown that amplification of the majority of reference genes used in quantifying the expression of mRNA varies according to experimental conditions. It is therefore necessary to carry out an initial
515:, which are undesirable potential by-products in PCR. However, fluorescent reporter probes do not prevent the inhibitory effect of the primer dimers, which may depress accumulation of the desired products in the reaction.
783:. Relative quantification is based on internal reference genes to determine fold-differences in expression of the target gene. The quantification is expressed as the change in expression levels of mRNA interpreted as
2590:
Yeh S.H. Tsai C.Y. Kao J.H. Liu C.J. Kuo T.J. Lin M.W. Huang W.L. Lu S.F. Jih J. Chen D.S. Others (2004). "Quantification and genotyping of hepatitis B virus in a single reaction by real-time PCR and melting …".
610:
electrophoresis is required it is only necessary to test those samples that real time PCR has shown to be doubtful and/or to ratify the results for samples that have tested positive for a specific determinant.
534:
with a laser. An increase in the product targeted by the reporter probe at each PCR cycle therefore causes a proportional increase in fluorescence due to the breakdown of the probe and release of the reporter.
475:
in PCR, increasing the fluorescence quantum yield of the dye. An increase in DNA product during PCR therefore leads to an increase in fluorescence intensity measured at each cycle. However, dsDNA dyes such as
954:. This quantification is carried out either with reverse transcription or without it, as occurs if the virus becomes integrated in the human genome at any point in its cycle, such as happens in the case of
186:
molecule during the PCR (i.e., in real time), not at its end, as in conventional PCR. Real-time PCR can be used quantitatively and semi-quantitatively (i.e., above/below a certain amount of DNA molecules).
795:
used, which can affect the efficiency of reverse transcription and therefore that of the whole PCR process. However, the most crucial aspect of the process is that the reference gene must be stable.
2124:
Xiu, Leshan; Binder, Raquel A.; Alarja, Natalie A.; Kochek, Kara; Coleman, Kristen K.; Than, Son T.; Bailey, Emily S.; Bui, Vuong N.; Toh, Teck-Hock; Erdman, Dean D.; Gray, Gregory C. (July 2020).
2941:
Kubista, M; Andrade, JM; Bengtsson, M; Forootan, A; Jonak, J; Lind, K; Sindelka, R; Sjoback, R; Sjogreen, B; Strombom, L; Stahlberg, A; Zoric, N (2006). "The real-time polymerase chain reaction".
913:
given its sensitivity and dynamic range in detecting DNA. Alternatives such as DNA or protein analysis are usually less sensitive. Specific primers are used that amplify not the transgene but the
821:
A number of statistical algorithms have been developed that can detect which gene or genes are most suitable for use under given conditions. Those like geNORM or BestKeeper can compare pairs or
699:
of RNA/DNA from a housekeeping gene in the same sample to normalize for variation in the amount and quality of RNA between different samples. This normalization procedure is commonly called the
557:
Fluorescence is detected and measured in a real-time PCR machine, and its geometric increase corresponding to exponential increase of the product is used to determine the quantification cycle
1489:
Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H (2006). "Quantitative real-time RT-PCR data analysis: current concepts and the novel "gene expression's CT difference" formula".
791:
of mRNA). Relative quantification is easier to carry out as it does not require a calibration curve as the amount of the studied gene is compared to the amount of a control reference gene.
897:
and other species, mixed in with the DNA of the host plant. Discrimination between the DNA of the pathogen and the plant is based on the amplification of ITS sequences, spacers located in
775:
by two common methods: relative quantification and absolute quantification. Absolute quantification gives the exact number of target DNA molecules by comparison with DNA standards using a
716:
provides reliable estimates of target DNA concentration in a sample under normal qPCR conditions, MAK2 does not reliably quantify target concentration for qPCR assays with competimeters.
348:. Northern blotting is often used to estimate the expression level of a gene by visualizing the abundance of its mRNA transcript in a sample. In this method, purified RNA is separated by
1334:"Real-time PCR based on SYBR-Green I fluorescence: An alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions"
1730:
522:
of fluorescence at the opposite end of the probe. The close proximity of the reporter to the quencher prevents detection of its fluorescence; breakdown of the probe by the 5' to 3'
942:. The degree of infection, quantified as the copies of the viral genome per unit of the patient's tissue, is relevant in many cases; for example, the probability that the type 1
511:
in multiplex assays for monitoring several target sequences in the same tube. The specificity of fluorescent reporter probes also prevents interference of measurements caused by
2779:
3123:
2506:
Holst-Jensen, Arne; Rønning, Sissel B.; Løvseth, Astrid; Berdal, Knut G. (2003). "PCR technology for screening and quantification of genetically modified organisms (GMOs)".
446:
carried out by the DNA polymerase. Due to the small size of the fragments the last step is usually omitted in this type of PCR as the enzyme is able to replicate the DNA
1785:
S. Dhanasekaran; T. Mark
Doherty; John Kenneth; TB Trials Study Group (March 2010). "Comparison of different standards for real-time PCR-based absolute quantification".
3302:
2400:
938:
and treatment. The use of qPCR allows both the quantification and genotyping (characterization of the strain, carried out using melting curves) of a virus such as the
3262:
230:
31:
901:
gene's coding area, which are characteristic for each taxon. Field-based versions of this technique have also been developed for identifying the same pathogen.
3290:
2013:
426:
with the capacity to illuminate each sample with a beam of light of at least one specified wavelength and detect the fluorescence emitted by the excited
2414:
Baldwin, B.G. (1992). "Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: An example from the
Compositaogy".
1283:"Development and Validation of an Externally Standardized Quantitative Insulin-like Growth Factor-1 RT-PCR Using LightCycler SYBR Green I Technology"
853:
has significantly improved the diagnosis of infectious diseases, and is deployed as a tool to detect newly emerging diseases, such as new strains of
642:
During the exponential amplification phase, the quantity of the target DNA template (amplicon) doubles every cycle. For example, a DNA sample whose C
393:
3320:
3116:
280:
In order to amplify small amounts of DNA, the same methodology is used as in conventional PCR using a DNA template, at least one pair of specific
107:
985:
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009).
79:
3243:
2628:"The Probability of in Vivo Reactivation of Herpes Simplex Virus Type 1 Increases with the Number of Latently Infected Neurons in the Ganglia"
1127:"Relative Expression Software Tool (REST©) for group wise comparison and statistical analysis of relative expression results in real-time PCR"
487:
In real-time PCR with dsDNA dyes the reaction is prepared as usual, with the addition of fluorescent dsDNA dye. Then the reaction is run in a
3092:
3018:
2328:
1316:
1060:
1035:
692:
684:
672:
656:
632:
558:
1534:"Development and evaluation of different normalization strategies for gene expression studies in Candida albicans biofilms by real-time PCR"
3331:
627:
of the signal noise above background. The number of cycles at which the fluorescence exceeds the threshold is called the threshold cycle (C
86:
2882:"Detection of specific polymerase chain reaction product by utilizing the 50 !30 exonuclease activity of Thermus aquaticus DNA polymerase"
736:. Uses of the technique in industry include the quantification of microbial load in foods or on vegetable matter, the detection of GMOs (
3296:
3280:
430:. The thermal cycler is also able to rapidly heat and chill samples, thereby taking advantage of the physicochemical properties of the
357:
3404:
3109:
2102:
93:
3314:
3268:
2843:
Higuchi, R.; Dollinger, G.; Walsh, P.S.; Griffith, R. (1992). "Simultaneous amplification and detection of specific DNA-sequences".
2794:
126:
463:
Real-time PCR technique can be classified by the chemistry used to detect the PCR product, specific or non-specific fluorochromes.
64:
57:
3274:
2097:
2970:
Higuchi, R.; Fockler, C.; Dollinger, G.; Watson, R. (1993). "Kinetic PCR: Real time monitoring of DNA amplification reactions".
764:
does not allow precise quantification. For example, over the 20–40 cycles of a typical PCR, the amount of DNA product reaches a
75:
3399:
737:
1332:
Ponchel F; Toomes C; Bransfield K; Leong F.T; Douglas S.H; Field S.L; Bell S.M; Combaret V; Puisieux A; Mighell A.J (2003).
3409:
3364:
1179:"Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes"
811:
530:
breaks the reporter-quencher proximity and thus allows unquenched emission of fluorescence, which can be detected after
195:
3153:
757:
584:
488:
349:
293:
2041:
3394:
370:
281:
211:
2294:
2273:
53:
3132:
540:
417:
266:
179:
100:
966:(CMV) which is seen in patients who are immunosuppressed following solid organ or bone marrow transplantation.
519:
2451:"Faster, Simpler, More-Specific Methods for Improved Molecular Detection of Phytophthora ramorum in the Field"
269:(PCR) is a common method for amplifying DNA; for RNA-based PCR the RNA sample is first reverse-transcribed to
3163:
1963:
Thellin, O; Zorzi, W; Lakaye, B; De Borman, B; Coumans, B; Henne, G; Grisar, T; Igout, A; Heinen, E (1999).
671:
is then used to determine the efficiency of amplification, which is 100% if a dilution of 1:2 results in a
2320:
341:
802:
using qualitative or semi-quantitative studies such as the visual examination of RNA gels, northern blot
484:). This can potentially interfere with, or prevent, accurate monitoring of the intended target sequence.
3233:
2810:"Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays"
2551:"… -Time Quantitative Polymerase Chain Reaction Methods for Four Genetically Modified Maize Varieties …"
2394:
963:
918:
788:
765:
274:
3194:
583:
Real-time PCR permits the identification of specific, amplified DNA fragments using analysis of their
3045:
2893:
2462:
2098:"How is the COVID-19 Virus Detected using real time reverse transcription–polymerase chain reaction?"
1639:
1443:
943:
889:
862:
337:
171:
987:"The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments"
3218:
3158:
1746:"The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression"
914:
850:
842:
729:
547:
452:
353:
285:
3351:
3223:
3213:
3208:
2995:
2911:
2868:
2531:
2375:
1608:
1514:
1177:
Vandesompele, J; De Preter, K; Pattyn, F; Poppe, B; Van Roy, N; De Paepe, A; Speleman, F (2002).
1107:
624:
1385:"Product Differentiation by Analysis of DNA Melting Curves during the Polymerase Chain Reaction"
2677:"MYC activation associated with the integration of HPV DNA at the MYC locus in genital tumours"
3198:
3088:
3071:
3014:
2987:
2958:
2929:
2860:
2831:
2757:
2739:
2698:
2657:
2608:
2572:
2523:
2488:
2431:
2324:
2255:
2204:
2155:
2078:
2033:
1994:
1945:
1910:
1859:
1841:
1802:
1767:
1703:
1667:
1600:
1565:
1506:
1471:
1407:
1365:
1312:
1263:
1210:
1156:
1099:
1056:
1031:
1008:
939:
799:
784:
776:
668:
620:
374:
270:
175:
2295:"rRT-PCR, a method to confirm Wuhan coronavirus case – Artificial Intelligence for Chemistry"
748:
Quantifying gene expression by traditional DNA detection methods is unreliable. Detection of
3256:
3061:
3053:
2979:
2950:
2919:
2901:
2852:
2821:
2747:
2729:
2688:
2647:
2639:
2600:
2562:
2515:
2478:
2470:
2423:
2367:
2245:
2235:
2194:
2186:
2145:
2137:
2068:
2025:
1984:
1976:
1937:
1900:
1890:
1849:
1833:
1794:
1757:
1744:
Overbergh, L.; Giulietti, A.; Valckx, D.; Decallonne, R.; Bouillon, R.; Mathieu, C. (2003).
1695:
1657:
1647:
1592:
1555:
1545:
1498:
1461:
1451:
1399:
1355:
1345:
1305:
1253:
1245:
1200:
1190:
1146:
1138:
1091:
998:
826:
780:
190:
Two common methods for the detection of PCR products in real-time PCR are (1) non-specific
3248:
3189:
959:
606:
442:
of the primers with the DNA template; the third, at between 68 and 72 °C, facilitates the
388:
The most commonly used normalizing genes are those that code for the following molecules:
382:
366:
362:
333:
289:
258:
250:
203:
191:
1095:
958:(human papillomavirus), where some of its variants are associated with the appearance of
3049:
2897:
2466:
1643:
1447:
467:
Non-specific detection: real-time PCR with double-stranded DNA-binding dyes as reporters
3378:
3228:
3203:
3101:
2805:
2483:
2450:
2250:
2223:
2199:
2174:
2150:
2125:
1854:
1821:
1762:
1745:
1662:
1627:
1560:
1533:
1466:
1431:
1282:
822:
733:
652:
527:
443:
435:
423:
378:
352:, transferred to a solid matrix (such as a nylon membrane), and probed with a specific
329:
301:
3066:
3029:
2752:
2717:
2652:
2627:
1980:
1905:
1878:
1583:
Nolan T, Hands RE, Bustin SA (2006). "Quantification of mRNA using real-time RT-PCR".
1360:
1333:
1258:
1229:
1205:
1178:
1151:
1126:
930:
Viruses can be present in humans due to direct infection or co-infections which makes
779:. It is therefore essential that the PCR of the sample and the standard have the same
3388:
3285:
2924:
2881:
2427:
2355:
2175:"Real-Time PCR in Clinical Microbiology: Applications for Routine Laboratory Testing"
1195:
898:
761:
753:
531:
405:
345:
3057:
2643:
2535:
2379:
1612:
1518:
518:
The method relies on a DNA-based probe with a fluorescent reporter at one end and a
3346:
3034:(ribulose-1,5-bisphosphate carboxylase/oxygenase) mRNA in diatoms and pelagophytes"
2999:
2872:
838:
810:
era, it is possible to carry out a more detailed estimate for many organisms using
803:
772:
574:
512:
507:
481:
431:
309:
217:
The
Minimum Information for Publication of Quantitative Real-Time PCR Experiments (
207:
2190:
2057:"Validation of housekeeping genes for normalizing RNA expression in real-time PCR"
1111:
768:
that is not directly correlated with the amount of target DNA in the initial PCR.
724:
There are numerous applications for quantitative polymerase chain reaction in the
455:
triphosphates (dNTPs) in the reaction and the bonding temperature of the primers.
1652:
1628:"A Mechanistic Model of PCR for Accurate Quantification of Quantitative PCR Data"
1456:
1384:
1003:
986:
480:
will bind to all dsDNA PCR products, including nonspecific PCR products (such as
3341:
1699:
858:
818:
sound methodological study in order to select the most suitable reference gene.
619:
real-time PCR relies on plotting fluorescence against the number of cycles on a
523:
427:
401:
297:
246:
241:
42:
3373:
2604:
2029:
1941:
1311:(3rd ed.). Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press.
3181:
2954:
2675:
Peter M. Rosty C. Couturier J. Radvanyi F. Teshima H. Sastre-garau X. (2006).
2519:
2371:
2141:
1798:
1502:
815:
725:
623:. A threshold for detection of DNA-based fluorescence is set 3–5 times of the
477:
313:
229:
be used for reverse transcription–qPCR. The acronym "RT-PCR" commonly denotes
143:
2743:
2567:
2550:
1879:"Validating internal controls for quantitative plant gene expression studies"
1845:
1249:
922:
control gene from the treated species that is only present as a single copy.
849:
and genetic abnormalities. The introduction of qualitative PCR assays to the
2906:
2734:
2274:"FDA-cleared RT-PCR Assays and Other Molecular Assays for Influenza Viruses"
2012:
Radonic, A; Thulke, S; Mackay, IM; Landt, O; Siegert, W; Nitsche, A (2004).
935:
931:
854:
648:
199:
3075:
2962:
2835:
2761:
2702:
2693:
2676:
2612:
2576:
2527:
2492:
2259:
2208:
2159:
2082:
2037:
1998:
1949:
1914:
1895:
1863:
1806:
1771:
1707:
1671:
1604:
1596:
1569:
1550:
1510:
1475:
1403:
1369:
1350:
1214:
1160:
1142:
1103:
1012:
499:
2991:
2933:
2864:
2826:
2809:
2661:
2435:
2055:
Dheda, K; Huggett, JF; Bustin, SA; Johnson, MA; Rook, G; Zumla, A (2004).
1432:"Robust Quantification of Polymerase Chain Reactions Using Global Fitting"
1411:
1267:
1051:
Watson, J D; Baker, T A; Bell, S P; Gann, A; Levine, M; Losick, R (2004).
150:
139:
3308:
2983:
2474:
1964:
1837:
951:
447:
2856:
2240:
798:
The selection of these reference genes was traditionally carried out in
2915:
2014:"Guideline for reference gene selection for quantitative real-time PCR"
397:
389:
2073:
2056:
233:
and not real-time PCR, but not all authors adhere to this convention.
1989:
947:
846:
807:
305:
2716:
Mackay, Ian M.; Arden, Katherine E.; Nitsche, Andreas (2002-03-15).
2126:"A RT-PCR assay for the detection of coronaviruses from four genera"
1532:
Nailis H, Coenye T, Van
Nieuwerburgh F, Deforce D, Nelis HJ (2006).
934:
difficult using classical techniques and can result in an incorrect
17:
3008:
691:
for an RNA or DNA from the gene of interest is subtracted from the
1026:
Logan, Julie; Edwards, Kirstin & Saunders, Nick, eds. (2009).
740:) and the quantification and genotyping of human viral pathogens.
664:
573:
498:
240:
149:
138:
30:
For reverse transcription polymerase chain reaction (RT-PCR), see
2880:
Holland, P.M.; Abramson, R.D.; Watson, R.; Gelfand, D.H. (1991).
1928:
McGettigan, Paul A (2013). "Transcriptomics in the RNA-seq era".
1230:"Optimization of the annealing temperature for DNA amplification
909:
qPCR using reverse transcription (RT-qPCR) can be used to detect
550:
stage of the PCR both probe and primers anneal to the DNA target.
749:
605:
Unlike conventional PCR, this method avoids the previous use of
218:
3105:
955:
910:
894:
472:
262:
183:
36:
651:
experiment with serial dilutions of DNA template to create a
631:) or, according to the MIQE guidelines, quantification cycle
312:
of the fluorophore after it has been excited at the required
2356:"Molecular Microbiology Methods For Environmental Diagnosis"
1820:
Bar, Tzachi; Kubista, Mikael; Tichopad, Ales (2011-10-19).
1965:"Housekeeping genes as internal standards: use and limits"
1430:
Carr, A. C.; Moore, S. D. (2012). Lucia, Alejandro (ed.).
837:
Diagnostic qualitative PCR is applied to rapidly detect
2549:
Brodmann P.D; Ilg E.C; Berthoud H; Herrmann A. (2002).
2224:"COVID-19: molecular and serological detection methods"
198:
with any double-stranded DNA and (2) sequence-specific
3309:
Co-amplification at lower denaturation temperature PCR
962:. Real-time PCR has also brought the quantization of
3362:
495:
Specific detection: fluorescent reporter probe method
336:. Older methods were used to measure mRNA abundance:
1055:(Fifth ed.). San Francisco: Benjamin Cummings.
471:
A DNA-binding dye binds to all double-stranded (ds)
365:
and many other factors. For this reason a number of
3330:
3242:
3180:
3140:
806:or semi-quantitative PCR (PCR mimics). Now, in the
182:(PCR). It monitors the amplification of a targeted
3085:Real-Time PCR: Current Technology and Applications
3010:Quantitative Real-time PCR in Applied Microbiology
2449:Tomlinson, J. A.; Barker, I.; Boonham, N. (2007).
2317:Quantitative Real-time PCR in Applied Microbiology
1304:
1028:Real-Time PCR: Current Technology and Applications
154:Melting curve produced at the end of real-time PCR
2780:"La PCR en temps réel: principes et applications"
946:reactivates is related to the number of infected
261:by turnover of gene transcripts (single stranded
3303:Multiplex ligation-dependent probe amplification
225:be used for quantitative real-time PCR and that
3263:Reverse transcription polymerase chain reaction
1877:Brunner, AM; Yakovlev, IA; Strauss, SH (2004).
1383:Ririe K.M; Rasmussen R.P; Wittwer C.T. (1997).
231:reverse transcription polymerase chain reaction
32:reverse transcription polymerase chain reaction
1172:
1170:
825:for a matrix of different reference genes and
214:of the probe with its complementary sequence.
3117:
3083:Logan J; Edwards K; Saunders N, eds. (2009).
210:reporter, which permits detection only after
8:
2399:: CS1 maint: multiple names: authors list (
980:
978:
146:fluorescence chart produced in real-time PCR
3291:Overlap extension polymerase chain reaction
1822:"Validation of kinetics similarity in qPCR"
1425:
1423:
1421:
1303:Sambrook, Joseph; Russel, David W. (2001).
1125:Pfaffl, MW; Horgan, GW; Dempfle, L (2002).
905:Detection of genetically modified organisms
221:) guidelines propose that the abbreviation
3124:
3110:
3102:
2348:
2346:
2344:
2342:
2340:
1626:Boggy G, Woolf PJ (2010). Ravasi T (ed.).
3065:
2923:
2905:
2825:
2751:
2733:
2692:
2651:
2566:
2482:
2249:
2239:
2198:
2149:
2072:
1988:
1904:
1894:
1853:
1761:
1723:"PEMF For Treatment Of Corneal Disorders"
1661:
1651:
1559:
1549:
1465:
1455:
1359:
1349:
1257:
1228:Rychlik W, Spencer WJ, Rhoads RE (1990).
1204:
1194:
1150:
1002:
127:Learn how and when to remove this message
27:Laboratory technique of molecular biology
3028:Wawrik, B; Paul, JH; Tabita, FR (2002).
1292:(3) – via gene-quantification.org.
1076:
1074:
1072:
579:from the expected amplification product.
394:glyceraldehyde-3-phosphate dehydrogenase
3369:
3321:Random amplification of polymorphic DNA
974:
3257:Quantitative polymerase chain reaction
2508:Analytical and Bioanalytical Chemistry
2455:Applied and Environmental Microbiology
2392:
1307:Molecular Cloning: A Laboratory Manual
926:Clinical quantification and genotyping
771:Real-time PCR can be used to quantify
546:As the reaction commences, during the
332:are modern methodologies for studying
63:Please improve this article by adding
2416:Molecular Phylogenetics and Evolution
841:that are diagnostic of, for example,
76:"Real-time polymerase chain reaction"
7:
2787:Reviews in Biology and Biotechnology
2222:Dhamad, AE; Abdal Rhida, MA (2020).
377:as their functions related to basic
3297:Multiplex polymerase chain reaction
3281:Touchdown polymerase chain reaction
1930:Current Opinion in Chemical Biology
543:), and the reporter probe is added.
160:real-time polymerase chain reaction
2103:International Atomic Energy Agency
1750:Journal of Biomolecular Techniques
1096:10.1023/b:bile.0000019559.84305.47
539:The PCR is prepared as usual (see
422:Real-time PCR is carried out in a
25:
3315:Digital polymerase chain reaction
3269:Inverse polymerase chain reaction
3030:"Real-time PCR quantification of
2096:Jawerth, Nicole (27 March 2020).
744:Quantification of gene expression
683:To quantify gene expression, the
3372:
3275:Nested polymerase chain reaction
1787:Journal of Immunological Methods
1196:10.1186/gb-2002-3-7-research0034
851:clinical microbiology laboratory
257:Cells in all organisms regulate
41:
3058:10.1128/aem.68.8.3771-3779.2002
2644:10.1128/JVI.72.8.6888-6892.1998
2360:Environmental Chemistry Letters
728:. It is commonly used for both
170:when used quantitatively) is a
738:genetically modified organisms
300:is added to this mixture in a
1:
2555:Journal of AOAC International
2191:10.1128/CMR.19.1.165-256.2006
2179:Clinical Microbiology Reviews
1981:10.1016/s0168-1656(99)00163-7
1053:Molecular Biology of the Gene
249:in order to detect levels of
65:secondary or tertiary sources
2778:Elyse; Houde, Alain (2002).
2428:10.1016/1055-7903(92)90030-K
2130:Journal of Clinical Virology
1653:10.1371/journal.pone.0012355
1457:10.1371/journal.pone.0037640
1004:10.1373/clinchem.2008.112797
383:constitutive gene expression
296:. A substance marked with a
294:thermo-stable DNA polymerase
2718:"Real-time PCR in virology"
2173:Espy, M.J. (January 2006).
1700:10.1016/j.ymeth.2012.08.011
882:Detection of phytopathogens
812:transcriptomic technologies
570:Fusion temperature analysis
350:agarose gel electrophoresis
3426:
3087:. Caister Academic Press.
3013:. Caister Academic Press.
2886:Proc. Natl. Acad. Sci. USA
2605:10.1016/j.jhep.2004.06.031
2030:10.1016/j.bbrc.2003.11.177
2018:Biochem Biophys Res Commun
1942:10.1016/j.cbpa.2012.12.008
1030:. Caister Academic Press.
415:
288:triphosphates, a suitable
29:
3405:Polymerase chain reaction
3133:Polymerase chain reaction
2955:10.1016/j.mam.2005.12.007
2793:(2): 2–11. Archived from
2520:10.1007/s00216-003-1767-7
2372:10.1007/s10311-016-0581-3
2142:10.1016/j.jcv.2020.104391
1799:10.1016/j.jim.2010.01.004
1503:10.1007/s00109-006-0097-6
893:, an oomycete that kills
418:Polymerase chain reaction
267:polymerase chain reaction
206:that are labelled with a
180:polymerase chain reaction
3038:Appl. Environ. Microbiol
1281:Pfaffl, Michael (2000).
781:amplification efficiency
663:with each dilution. The
489:real-time PCR instrument
381:survival normally imply
319:relative gene expression
2907:10.1073/pnas.88.16.7276
2315:Filion, M, ed. (2012).
1727:lemuriatechnologies.com
1392:Analytical Biochemistry
459:Chemical classification
2722:Nucleic Acids Research
2694:10.1038/sj.onc.1209625
2568:10.1093/jaoac/85.3.646
2321:Caister Academic Press
1896:10.1186/1471-2229-4-14
1826:Nucleic Acids Research
1597:10.1038/nprot.2006.236
1551:10.1186/1471-2199-7-25
1404:10.1006/abio.1996.9916
1351:10.1186/1472-6750-3-18
1250:10.1093/nar/18.21.6409
580:
504:
369:systems (often called
342:RNase protection assay
254:
155:
147:
52:relies excessively on
3400:Laboratory techniques
2827:10.1677/jme.0.0250169
2735:10.1093/nar/30.6.1292
2626:Sawtell N.M. (1998).
2593:Journal of Hepatology
1084:Biotechnology Letters
964:human cytomegalovirus
789:reverse transcription
756:or PCR products on a
577:
502:
371:normalization methods
328:Quantitative PCR and
275:reverse transcriptase
244:
153:
142:
3410:Real-time technology
2984:10.1038/nbt0993-1026
2475:10.1128/AEM.00161-07
1143:10.1093/nar/30.9.e36
944:herpes simplex virus
890:Phytophthora ramorum
869:Microbiological uses
787:(cDNA, generated by
338:differential display
172:laboratory technique
3050:2002ApEnM..68.3771W
3007:Filion, M. (2012).
2898:1991PNAS...88.7276H
2857:10.1038/nbt0492-413
2632:Journal of Virology
2467:2007ApEnM..73.4040T
2241:10.7717/peerj.10180
1644:2010PLoSO...512355B
1448:2012PLoSO...737640C
843:infectious diseases
585:melting temperature
453:deoxyribonucleotide
286:deoxyribonucleotide
245:Real time PCR uses
1838:10.1093/nar/gkr778
991:Clinical Chemistry
625:standard deviation
581:
505:
375:housekeeping genes
361:of DNA integrity,
308:for measuring the
255:
156:
148:
3395:Molecular biology
3360:
3359:
3094:978-1-904455-39-4
3020:978-1-908230-01-0
2892:(16): 7276–7280.
2687:(44): 5985–5993.
2461:(12): 4040–4047.
2330:978-1-908230-01-0
2297:. 24 January 2020
2074:10.2144/04371RR03
1318:978-0-87969-576-7
1244:(21): 6409–6412.
1238:Nucleic Acids Res
1131:Nucleic Acids Res
1062:978-0-321-22368-5
1037:978-1-904455-39-4
940:hepatitis B virus
800:molecular biology
785:complementary DNA
777:calibration curve
669:linear regression
655:of the change in
621:logarithmic scale
565:in each reaction.
363:enzyme efficiency
271:complementary DNA
176:molecular biology
137:
136:
129:
111:
16:(Redirected from
3417:
3377:
3376:
3368:
3170:Final elongation
3126:
3119:
3112:
3103:
3098:
3079:
3069:
3044:(8): 3771–3779.
3024:
3003:
2978:(9): 1026–1030.
2966:
2943:Mol. Aspects Med
2937:
2927:
2909:
2876:
2839:
2829:
2814:J Mol Endocrinol
2801:
2799:
2784:
2766:
2765:
2755:
2737:
2728:(6): 1292–1305.
2713:
2707:
2706:
2696:
2672:
2666:
2665:
2655:
2638:(8): 6888–6892.
2623:
2617:
2616:
2587:
2581:
2580:
2570:
2546:
2540:
2539:
2503:
2497:
2496:
2486:
2446:
2440:
2439:
2411:
2405:
2404:
2398:
2390:
2388:
2386:
2350:
2335:
2334:
2312:
2306:
2305:
2303:
2302:
2291:
2285:
2284:
2278:
2270:
2264:
2263:
2253:
2243:
2219:
2213:
2212:
2202:
2170:
2164:
2163:
2153:
2121:
2115:
2114:
2112:
2110:
2093:
2087:
2086:
2076:
2052:
2046:
2045:
2040:. Archived from
2009:
2003:
2002:
1992:
1975:(2–3): 197–200.
1960:
1954:
1953:
1925:
1919:
1918:
1908:
1898:
1874:
1868:
1867:
1857:
1832:(4): 1395–1406.
1817:
1811:
1810:
1782:
1776:
1775:
1765:
1741:
1735:
1734:
1729:. Archived from
1721:Bruce Gelerter.
1718:
1712:
1711:
1682:
1676:
1675:
1665:
1655:
1623:
1617:
1616:
1591:(3): 1559–1582.
1580:
1574:
1573:
1563:
1553:
1529:
1523:
1522:
1486:
1480:
1479:
1469:
1459:
1427:
1416:
1415:
1389:
1380:
1374:
1373:
1363:
1353:
1329:
1323:
1322:
1310:
1300:
1294:
1293:
1287:
1278:
1272:
1271:
1261:
1225:
1219:
1218:
1208:
1198:
1174:
1165:
1164:
1154:
1122:
1116:
1115:
1078:
1067:
1066:
1048:
1042:
1041:
1023:
1017:
1016:
1006:
982:
863:diagnostic tests
526:activity of the
412:Basic principles
354:DNA or RNA probe
323:mRNA copy number
204:oligonucleotides
192:fluorescent dyes
132:
125:
121:
118:
112:
110:
69:
45:
37:
21:
3425:
3424:
3420:
3419:
3418:
3416:
3415:
3414:
3385:
3384:
3383:
3371:
3363:
3361:
3356:
3334:
3326:
3299:(multiplex PCR)
3246:
3238:
3176:
3136:
3130:
3095:
3082:
3027:
3021:
3006:
2969:
2949:(2–3): 95–125.
2940:
2879:
2842:
2804:
2797:
2782:
2777:
2774:
2769:
2715:
2714:
2710:
2674:
2673:
2669:
2625:
2624:
2620:
2589:
2588:
2584:
2548:
2547:
2543:
2505:
2504:
2500:
2448:
2447:
2443:
2413:
2412:
2408:
2391:
2384:
2382:
2352:
2351:
2338:
2331:
2314:
2313:
2309:
2300:
2298:
2293:
2292:
2288:
2276:
2272:
2271:
2267:
2221:
2220:
2216:
2172:
2171:
2167:
2123:
2122:
2118:
2108:
2106:
2095:
2094:
2090:
2054:
2053:
2049:
2011:
2010:
2006:
1962:
1961:
1957:
1927:
1926:
1922:
1876:
1875:
1871:
1819:
1818:
1814:
1784:
1783:
1779:
1743:
1742:
1738:
1720:
1719:
1715:
1684:
1683:
1679:
1625:
1624:
1620:
1582:
1581:
1577:
1531:
1530:
1526:
1497:(11): 901–910.
1488:
1487:
1483:
1429:
1428:
1419:
1387:
1382:
1381:
1377:
1331:
1330:
1326:
1319:
1302:
1301:
1297:
1285:
1280:
1279:
1275:
1227:
1226:
1222:
1176:
1175:
1168:
1124:
1123:
1119:
1080:
1079:
1070:
1063:
1050:
1049:
1045:
1038:
1025:
1024:
1020:
984:
983:
976:
972:
960:cervical cancer
928:
907:
884:
871:
835:
833:Diagnostic uses
823:geometric means
746:
722:
704:
696:
688:
676:
660:
645:
636:
630:
616:
607:electrophoresis
592:
572:
562:
497:
469:
461:
420:
414:
367:standardization
334:gene expression
290:buffer solution
259:gene expression
251:gene expression
239:
133:
122:
116:
113:
70:
68:
62:
58:primary sources
46:
35:
28:
23:
22:
15:
12:
11:
5:
3423:
3421:
3413:
3412:
3407:
3402:
3397:
3387:
3386:
3382:
3381:
3358:
3357:
3355:
3354:
3349:
3344:
3338:
3336:
3328:
3327:
3325:
3324:
3318:
3312:
3306:
3300:
3294:
3288:
3283:
3278:
3272:
3266:
3260:
3253:
3251:
3240:
3239:
3237:
3236:
3231:
3226:
3221:
3216:
3211:
3206:
3201:
3192:
3186:
3184:
3178:
3177:
3175:
3174:
3171:
3168:
3167:
3166:
3161:
3156:
3148:
3147:Initialization
3144:
3142:
3138:
3137:
3131:
3129:
3128:
3121:
3114:
3106:
3100:
3099:
3093:
3080:
3025:
3019:
3004:
2967:
2938:
2877:
2851:(4): 413–417.
2845:Bio-Technology
2840:
2820:(2): 169–193.
2802:
2800:on 2009-06-12.
2773:
2770:
2768:
2767:
2708:
2667:
2618:
2599:(4): 659–666.
2582:
2561:(3): 646–653.
2541:
2514:(8): 985–993.
2498:
2441:
2406:
2366:(4): 423–441.
2336:
2329:
2307:
2286:
2265:
2214:
2185:(3): 165–256.
2165:
2116:
2088:
2067:(1): 112–119.
2047:
2044:on 2013-08-02.
2024:(4): 856–862.
2004:
1955:
1920:
1883:BMC Plant Biol
1869:
1812:
1793:(1–2): 34–39.
1777:
1736:
1733:on 2014-06-09.
1713:
1677:
1618:
1575:
1524:
1481:
1417:
1398:(2): 154–160.
1375:
1338:BMC Biotechnol
1324:
1317:
1295:
1273:
1220:
1183:Genome Biology
1166:
1117:
1068:
1061:
1043:
1036:
1018:
997:(4): 611–622.
973:
971:
968:
927:
924:
906:
903:
883:
880:
870:
867:
834:
831:
745:
742:
734:basic research
721:
718:
702:
694:
686:
674:
658:
653:standard curve
643:
634:
628:
615:
612:
590:
571:
568:
567:
566:
560:
555:
551:
544:
528:Taq polymerase
496:
493:
468:
465:
460:
457:
444:polymerization
436:DNA polymerase
424:thermal cycler
416:Main article:
413:
410:
406:ribosomal RNAs
330:DNA microarray
304:that contains
302:thermal cycler
238:
235:
202:consisting of
135:
134:
49:
47:
40:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
3422:
3411:
3408:
3406:
3403:
3401:
3398:
3396:
3393:
3392:
3390:
3380:
3375:
3370:
3366:
3353:
3350:
3348:
3345:
3343:
3340:
3339:
3337:
3333:
3329:
3322:
3319:
3316:
3313:
3310:
3307:
3304:
3301:
3298:
3295:
3292:
3289:
3287:
3286:Hot start PCR
3284:
3282:
3279:
3276:
3273:
3271:(inverse PCR)
3270:
3267:
3264:
3261:
3258:
3255:
3254:
3252:
3250:
3245:
3241:
3235:
3232:
3230:
3227:
3225:
3222:
3220:
3217:
3215:
3212:
3210:
3207:
3205:
3202:
3200:
3196:
3193:
3191:
3188:
3187:
3185:
3183:
3179:
3172:
3169:
3165:
3162:
3160:
3157:
3155:
3152:
3151:
3149:
3146:
3145:
3143:
3139:
3134:
3127:
3122:
3120:
3115:
3113:
3108:
3107:
3104:
3096:
3090:
3086:
3081:
3077:
3073:
3068:
3063:
3059:
3055:
3051:
3047:
3043:
3039:
3035:
3033:
3026:
3022:
3016:
3012:
3011:
3005:
3001:
2997:
2993:
2989:
2985:
2981:
2977:
2973:
2972:Biotechnology
2968:
2964:
2960:
2956:
2952:
2948:
2944:
2939:
2935:
2931:
2926:
2921:
2917:
2913:
2908:
2903:
2899:
2895:
2891:
2887:
2883:
2878:
2874:
2870:
2866:
2862:
2858:
2854:
2850:
2846:
2841:
2837:
2833:
2828:
2823:
2819:
2815:
2811:
2807:
2803:
2796:
2792:
2788:
2781:
2776:
2775:
2771:
2763:
2759:
2754:
2749:
2745:
2741:
2736:
2731:
2727:
2723:
2719:
2712:
2709:
2704:
2700:
2695:
2690:
2686:
2682:
2678:
2671:
2668:
2663:
2659:
2654:
2649:
2645:
2641:
2637:
2633:
2629:
2622:
2619:
2614:
2610:
2606:
2602:
2598:
2594:
2586:
2583:
2578:
2574:
2569:
2564:
2560:
2556:
2552:
2545:
2542:
2537:
2533:
2529:
2525:
2521:
2517:
2513:
2509:
2502:
2499:
2494:
2490:
2485:
2480:
2476:
2472:
2468:
2464:
2460:
2456:
2452:
2445:
2442:
2437:
2433:
2429:
2425:
2421:
2417:
2410:
2407:
2402:
2396:
2381:
2377:
2373:
2369:
2365:
2361:
2357:
2349:
2347:
2345:
2343:
2341:
2337:
2332:
2326:
2322:
2318:
2311:
2308:
2296:
2290:
2287:
2282:
2275:
2269:
2266:
2261:
2257:
2252:
2247:
2242:
2237:
2233:
2229:
2225:
2218:
2215:
2210:
2206:
2201:
2196:
2192:
2188:
2184:
2180:
2176:
2169:
2166:
2161:
2157:
2152:
2147:
2143:
2139:
2135:
2131:
2127:
2120:
2117:
2105:
2104:
2099:
2092:
2089:
2084:
2080:
2075:
2070:
2066:
2062:
2061:BioTechniques
2058:
2051:
2048:
2043:
2039:
2035:
2031:
2027:
2023:
2019:
2015:
2008:
2005:
2000:
1996:
1991:
1986:
1982:
1978:
1974:
1970:
1966:
1959:
1956:
1951:
1947:
1943:
1939:
1935:
1931:
1924:
1921:
1916:
1912:
1907:
1902:
1897:
1892:
1888:
1884:
1880:
1873:
1870:
1865:
1861:
1856:
1851:
1847:
1843:
1839:
1835:
1831:
1827:
1823:
1816:
1813:
1808:
1804:
1800:
1796:
1792:
1788:
1781:
1778:
1773:
1769:
1764:
1759:
1755:
1751:
1747:
1740:
1737:
1732:
1728:
1724:
1717:
1714:
1709:
1705:
1701:
1697:
1693:
1689:
1681:
1678:
1673:
1669:
1664:
1659:
1654:
1649:
1645:
1641:
1638:(8): e12355.
1637:
1633:
1629:
1622:
1619:
1614:
1610:
1606:
1602:
1598:
1594:
1590:
1586:
1579:
1576:
1571:
1567:
1562:
1557:
1552:
1547:
1543:
1539:
1538:BMC Mol. Biol
1535:
1528:
1525:
1520:
1516:
1512:
1508:
1504:
1500:
1496:
1492:
1485:
1482:
1477:
1473:
1468:
1463:
1458:
1453:
1449:
1445:
1442:(5): e37640.
1441:
1437:
1433:
1426:
1424:
1422:
1418:
1413:
1409:
1405:
1401:
1397:
1393:
1386:
1379:
1376:
1371:
1367:
1362:
1357:
1352:
1347:
1343:
1339:
1335:
1328:
1325:
1320:
1314:
1309:
1308:
1299:
1296:
1291:
1284:
1277:
1274:
1269:
1265:
1260:
1255:
1251:
1247:
1243:
1239:
1235:
1233:
1224:
1221:
1216:
1212:
1207:
1202:
1197:
1192:
1188:
1184:
1180:
1173:
1171:
1167:
1162:
1158:
1153:
1148:
1144:
1140:
1136:
1132:
1128:
1121:
1118:
1113:
1109:
1105:
1101:
1097:
1093:
1090:(6): 509–15.
1089:
1085:
1077:
1075:
1073:
1069:
1064:
1058:
1054:
1047:
1044:
1039:
1033:
1029:
1022:
1019:
1014:
1010:
1005:
1000:
996:
992:
988:
981:
979:
975:
969:
967:
965:
961:
957:
953:
949:
945:
941:
937:
933:
925:
923:
920:
916:
912:
904:
902:
900:
899:ribosomal RNA
896:
892:
891:
881:
879:
875:
868:
866:
864:
860:
856:
852:
848:
844:
840:
839:nucleic acids
832:
830:
828:
824:
819:
817:
816:statistically
813:
809:
805:
801:
796:
792:
790:
786:
782:
778:
774:
773:nucleic acids
769:
767:
763:
762:Southern blot
759:
755:
754:northern blot
751:
743:
741:
739:
735:
731:
727:
719:
717:
713:
709:
706:
698:
690:
681:
678:
670:
666:
662:
654:
650:
640:
638:
626:
622:
613:
611:
608:
603:
601:
597:
594:value, from m
593:
587:(also called
586:
576:
569:
564:
556:
554:fluorescence.
552:
549:
545:
542:
538:
537:
536:
533:
529:
525:
521:
516:
514:
513:primer dimers
509:
501:
494:
492:
490:
485:
483:
479:
474:
466:
464:
458:
456:
454:
449:
445:
439:
437:
433:
432:nucleic acids
429:
425:
419:
411:
409:
407:
403:
399:
395:
391:
386:
384:
380:
376:
372:
368:
364:
359:
358:complementary
355:
351:
347:
346:northern blot
343:
339:
335:
331:
326:
324:
320:
315:
311:
307:
303:
299:
295:
291:
287:
283:
278:
276:
272:
268:
264:
260:
252:
248:
243:
236:
234:
232:
228:
224:
220:
215:
213:
212:hybridization
209:
205:
201:
197:
193:
188:
185:
181:
178:based on the
177:
173:
169:
165:
164:real-time PCR
161:
152:
145:
141:
131:
128:
120:
109:
106:
102:
99:
95:
92:
88:
85:
81:
78: –
77:
73:
72:Find sources:
66:
60:
59:
55:
50:This article
48:
44:
39:
38:
33:
19:
3347:Kjell Kleppe
3277:(nested PCR)
3244:Optimization
3154:Denaturation
3084:
3041:
3037:
3031:
3009:
2975:
2971:
2946:
2942:
2889:
2885:
2848:
2844:
2817:
2813:
2795:the original
2790:
2786:
2772:Bibliography
2725:
2721:
2711:
2684:
2680:
2670:
2635:
2631:
2621:
2596:
2592:
2585:
2558:
2554:
2544:
2511:
2507:
2501:
2458:
2454:
2444:
2419:
2415:
2409:
2395:cite journal
2383:. Retrieved
2363:
2359:
2316:
2310:
2299:. Retrieved
2289:
2280:
2268:
2231:
2227:
2217:
2182:
2178:
2168:
2133:
2129:
2119:
2107:. Retrieved
2101:
2091:
2064:
2060:
2050:
2042:the original
2021:
2017:
2007:
1972:
1969:J Biotechnol
1968:
1958:
1933:
1929:
1923:
1886:
1882:
1872:
1829:
1825:
1815:
1790:
1786:
1780:
1756:(1): 33–43.
1753:
1749:
1739:
1731:the original
1726:
1716:
1694:(1): 32–46.
1691:
1687:
1680:
1635:
1631:
1621:
1588:
1584:
1578:
1541:
1537:
1527:
1494:
1490:
1484:
1439:
1435:
1395:
1391:
1378:
1341:
1337:
1327:
1306:
1298:
1289:
1276:
1241:
1237:
1231:
1223:
1186:
1182:
1134:
1130:
1120:
1087:
1083:
1052:
1046:
1027:
1021:
994:
990:
929:
908:
888:
885:
876:
872:
836:
820:
804:densitometry
797:
793:
770:
747:
723:
720:Applications
714:
710:
700:
682:
641:
617:
604:
599:
595:
588:
582:
517:
506:
486:
482:primer dimer
470:
462:
440:
421:
387:
327:
322:
318:
310:fluorescence
279:
273:(cDNA) with
256:
247:fluorophores
226:
222:
216:
189:
167:
163:
159:
157:
123:
114:
104:
97:
90:
83:
71:
51:
3352:Alice Chien
3342:Kary Mullis
2422:(1): 3–16.
2109:16 February
1936:(1): 4–11.
1585:Nat. Protoc
1189:(7): 1–12.
859:coronavirus
524:exonuclease
508:Fluorescent
428:fluorophore
402:cyclophilin
298:fluorophore
208:fluorescent
196:intercalate
3389:Categories
3335:and people
3311:(COLD-PCR)
3182:Polymerase
3173:Final hold
3164:Elongation
3135:techniques
2806:Bustin, SA
2301:2020-01-26
2234:: e10180.
2136:: 104391.
1290:Biochemica
1137:(9): e36.
970:References
919:terminator
730:diagnostic
726:laboratory
600:emperature
532:excitation
478:SYBR Green
314:wavelength
237:Background
200:DNA probes
144:SYBR Green
87:newspapers
54:references
3159:Annealing
3141:Procedure
2744:0305-1048
1990:2268/3661
1846:0305-1048
1544:(1): 25.
1491:J Mol Med
936:prognosis
932:diagnosis
649:titration
548:annealing
117:July 2018
3293:(OE-PCR)
3265:(RT-PCR)
3249:variants
3076:12147471
2963:16460794
2836:11013345
2808:(2000).
2762:11884626
2703:16682952
2681:Oncogene
2613:15464248
2577:12083257
2536:43681211
2528:12733008
2493:17449689
2380:88827291
2260:33083156
2209:16418529
2160:32403008
2083:15283208
2038:14706621
1999:10617337
1950:23290152
1915:15317655
1864:22013160
1807:20109462
1772:12901609
1708:22975077
1672:20814578
1632:PLOS ONE
1613:10108148
1605:17406449
1570:16889665
1519:10427299
1511:16972087
1476:22701526
1436:PLOS ONE
1370:14552656
1232:in vitro
1215:12184808
1161:11972351
1104:15127793
1013:19246619
915:promoter
614:Modeling
520:quencher
448:amplicon
379:cellular
356:that is
3379:Biology
3332:History
3317:(dePCR)
3046:Bibcode
3000:5714001
2992:7764001
2934:1871133
2916:2357665
2894:Bibcode
2873:1684150
2865:1368485
2662:9658140
2484:1932743
2463:Bibcode
2436:1342921
2281:cdc.gov
2251:7547594
2200:1360278
2151:7192118
1855:3287174
1763:2279895
1688:Methods
1663:2930010
1640:Bibcode
1561:1557526
1467:3365123
1444:Bibcode
1412:9056205
1268:2243783
952:ganglia
950:in the
948:neurons
827:tissues
766:plateau
705:-method
667:of the
398:albumin
390:tubulin
306:sensors
282:primers
227:RT-qPCR
101:scholar
3365:Portal
3323:(RAPD)
3305:(MLPA)
3259:(qPCR)
3190:Klenow
3150:Cycle
3091:
3074:
3067:123995
3064:
3017:
2998:
2990:
2961:
2932:
2922:
2914:
2871:
2863:
2834:
2760:
2753:101343
2750:
2742:
2701:
2660:
2653:109900
2650:
2611:
2575:
2534:
2526:
2491:
2481:
2434:
2385:11 May
2378:
2327:
2258:
2248:
2207:
2197:
2158:
2148:
2081:
2036:
1997:
1948:
1913:
1906:515301
1903:
1889:: 14.
1862:
1852:
1844:
1805:
1770:
1760:
1706:
1670:
1660:
1611:
1603:
1568:
1558:
1517:
1509:
1474:
1464:
1410:
1368:
1361:270040
1358:
1344:: 18.
1315:
1266:
1259:332522
1256:
1213:
1206:126239
1203:
1159:
1152:113859
1149:
1112:977404
1110:
1102:
1059:
1034:
1011:
847:cancer
808:genome
596:elting
404:, and
292:and a
103:
96:
89:
82:
74:
2996:S2CID
2925:52277
2912:JSTOR
2869:S2CID
2798:(PDF)
2783:(PDF)
2532:S2CID
2376:S2CID
2277:(PDF)
2228:PeerJ
1609:S2CID
1515:S2CID
1388:(PDF)
1286:(PDF)
1108:S2CID
861:, in
752:on a
665:slope
194:that
166:, or
108:JSTOR
94:books
3247:and
3219:Vent
3197:and
3089:ISBN
3072:PMID
3032:rbcL
3015:ISBN
2988:PMID
2959:PMID
2930:PMID
2861:PMID
2832:PMID
2758:PMID
2740:ISSN
2699:PMID
2658:PMID
2609:PMID
2573:PMID
2524:PMID
2489:PMID
2432:PMID
2401:link
2387:2020
2325:ISBN
2256:PMID
2205:PMID
2156:PMID
2111:2023
2079:PMID
2034:PMID
1995:PMID
1946:PMID
1911:PMID
1860:PMID
1842:ISSN
1803:PMID
1768:PMID
1704:PMID
1668:PMID
1601:PMID
1566:PMID
1507:PMID
1472:PMID
1408:PMID
1366:PMID
1313:ISBN
1264:PMID
1211:PMID
1157:PMID
1100:PMID
1057:ISBN
1032:ISBN
1009:PMID
911:GMOs
895:oaks
857:and
750:mRNA
732:and
434:and
344:and
321:(or
223:qPCR
219:MIQE
168:qPCR
80:news
18:QPCR
3234:Tfu
3229:Tli
3224:Pwo
3214:Pfu
3209:Tth
3204:Taq
3062:PMC
3054:doi
2980:doi
2951:doi
2920:PMC
2902:doi
2853:doi
2822:doi
2748:PMC
2730:doi
2689:doi
2648:PMC
2640:doi
2601:doi
2563:doi
2516:doi
2512:375
2479:PMC
2471:doi
2424:doi
2368:doi
2246:PMC
2236:doi
2195:PMC
2187:doi
2146:PMC
2138:doi
2134:128
2069:doi
2026:doi
2022:313
1985:hdl
1977:doi
1938:doi
1901:PMC
1891:doi
1850:PMC
1834:doi
1795:doi
1791:354
1758:PMC
1696:doi
1658:PMC
1648:doi
1593:doi
1556:PMC
1546:doi
1499:doi
1462:PMC
1452:doi
1400:doi
1396:245
1356:PMC
1346:doi
1254:PMC
1246:doi
1201:PMC
1191:doi
1147:PMC
1139:doi
1092:doi
999:doi
956:HPV
855:flu
760:or
758:gel
541:PCR
473:DNA
263:RNA
184:DNA
174:of
56:to
3391::
3199:T7
3195:T4
3070:.
3060:.
3052:.
3042:68
3040:.
3036:.
2994:.
2986:.
2976:11
2974:.
2957:.
2947:27
2945:.
2928:.
2918:.
2910:.
2900:.
2890:88
2888:.
2884:.
2867:.
2859:.
2849:10
2847:.
2830:.
2818:25
2816:.
2812:.
2789:.
2785:.
2756:.
2746:.
2738:.
2726:30
2724:.
2720:.
2697:.
2685:25
2683:.
2679:.
2656:.
2646:.
2636:72
2634:.
2630:.
2607:.
2597:41
2595:.
2571:.
2559:85
2557:.
2553:.
2530:.
2522:.
2510:.
2487:.
2477:.
2469:.
2459:73
2457:.
2453:.
2430:.
2418:.
2397:}}
2393:{{
2374:.
2364:14
2362:.
2358:.
2339:^
2323:.
2319:.
2279:.
2254:.
2244:.
2230:.
2226:.
2203:.
2193:.
2183:19
2181:.
2177:.
2154:.
2144:.
2132:.
2128:.
2100:.
2077:.
2065:37
2063:.
2059:.
2032:.
2020:.
2016:.
1993:.
1983:.
1973:75
1971:.
1967:.
1944:.
1934:17
1932:.
1909:.
1899:.
1885:.
1881:.
1858:.
1848:.
1840:.
1830:40
1828:.
1824:.
1801:.
1789:.
1766:.
1754:14
1752:.
1748:.
1725:.
1702:.
1692:59
1690:.
1666:.
1656:.
1646:.
1634:.
1630:.
1607:.
1599:.
1587:.
1564:.
1554:.
1540:.
1536:.
1513:.
1505:.
1495:84
1493:.
1470:.
1460:.
1450:.
1438:.
1434:.
1420:^
1406:.
1394:.
1390:.
1364:.
1354:.
1340:.
1336:.
1288:.
1262:.
1252:.
1242:18
1240:.
1236:.
1209:.
1199:.
1185:.
1181:.
1169:^
1155:.
1145:.
1135:30
1133:.
1129:.
1106:.
1098:.
1088:26
1086:.
1071:^
1007:.
995:55
993:.
989:.
977:^
917:,
865:.
845:,
829:.
701:ΔC
693:(C
685:(C
673:(C
657:(C
639:.
633:(C
559:(C
438:.
408:.
400:,
396:,
392:,
340:,
284:,
277:.
158:A
67:.
3367::
3125:e
3118:t
3111:v
3097:.
3078:.
3056::
3048::
3023:.
3002:.
2982::
2965:.
2953::
2936:.
2904::
2896::
2875:.
2855::
2838:.
2824::
2791:2
2764:.
2732::
2705:.
2691::
2664:.
2642::
2615:.
2603::
2579:.
2565::
2538:.
2518::
2495:.
2473::
2465::
2438:.
2426::
2420:1
2403:)
2389:.
2370::
2333:.
2304:.
2283:.
2262:.
2238::
2232:8
2211:.
2189::
2162:.
2140::
2113:.
2085:.
2071::
2028::
2001:.
1987::
1979::
1952:.
1940::
1917:.
1893::
1887:4
1866:.
1836::
1809:.
1797::
1774:.
1710:.
1698::
1674:.
1650::
1642::
1636:5
1615:.
1595::
1589:1
1572:.
1548::
1542:7
1521:.
1501::
1478:.
1454::
1446::
1440:7
1414:.
1402::
1372:.
1348::
1342:3
1321:.
1270:.
1248::
1234:"
1217:.
1193::
1187:3
1163:.
1141::
1114:.
1094::
1065:.
1040:.
1015:.
1001::
703:t
697:)
695:q
689:)
687:q
677:)
675:q
661:)
659:q
644:q
637:)
635:q
629:t
598:t
591:m
589:T
563:)
561:q
253:.
162:(
130:)
124:(
119:)
115:(
105:·
98:·
91:·
84:·
61:.
34:.
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.