340:. Regulation of translation can impact the global rate of protein synthesis which is closely coupled to the metabolic and proliferative state of a cell. To delve deeper into this intricate process, scientists typically use a technique known as ribosome profiling. This method enables researchers to take a snapshot of the translatome, showing which parts of the mRNA are being translated into proteins by ribosomes at a given time. Ribosome profiling provides valuable insights into translation dynamics, revealing the complex interplay between gene sequence, mRNA structure, and translation regulation. Expanding on this concept, a more recent development is single-cell ribosome profiling, a technique that allows us to study the translation process at the resolution of individual cells. Single-cell ribosome profiling has the potential to shed light on the heterogeneous nature of cells, leading to a more nuanced understanding of how translation regulation can impact cell behavior, metabolic state, and responsiveness to various stimuli or conditions.
268:, and the A site is ready to receive an aminoacyl-tRNA. During chain elongation, each additional amino acid is added to the nascent polypeptide chain in a three-step microcycle. The steps in this microcycle are (1) positioning the correct aminoacyl-tRNA in the A site of the ribosome, which is brought into that site by eEF1, (2) forming the peptide bond, and (3) shifting the mRNA by one codon relative to the ribosome with the help of eEF2. Unlike bacteria, in which translation initiation occurs as soon as the 5' end of an mRNA is synthesized, in eukaryotes, such tight coupling between transcription and translation is not possible because transcription and translation are carried out in separate compartments of the cell (the
58:
312:, eRF1, that recognizes all three stop codons. Upon termination, the ribosome is disassembled and the completed polypeptide is released. eRF3 is a ribosome-dependent GTPase that helps eRF1 release the completed polypeptide. The human genome encodes a few genes whose mRNA stop codon are surprisingly leaky: In these genes, termination of translation is inefficient due to special RNA bases in the vicinity of the stop codon. Leaky termination in these genes leads to
75:
240:(IRES). Unlike cap-dependent translation, cap-independent translation does not require a 5' cap to initiate scanning from the 5' end of the mRNA until the start codon. The ribosome can localize to the start site by direct binding, initiation factors, and/or ITAFs (IRES trans-acting factors) bypassing the need to scan the entire
65:
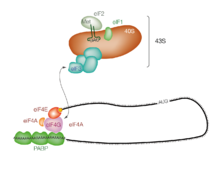
Translation initiation is the process by which the ribosome and its associated factors bind to an mRNA and are assembled at the start codon. This process is defined as either cap-dependent, in which the ribosome binds initially at the 5' cap and then travels to the stop codon, or as cap-independent,
128:
is the cap-binding protein. Binding of the cap by eIF4E is often considered the rate-limiting step of cap-dependent initiation, and the concentration of eIF4E is a regulatory nexus of translational control. Certain viruses cleave a portion of eIF4G that binds eIF4E, thus preventing cap-dependent
215:
While protein synthesis is globally regulated by modulating the expression of key initiation factors as well as the number of ribosomes, individual mRNAs can have different translation rates due to the presence of regulatory sequence elements. This has been shown to be important in a variety of
292:, which can trigger endonucleolytic attack of the tRNA, a process termed mRNA no-go decay. Ribosomal pausing also aids co-translational folding of the nascent polypeptide on the ribosome, and delays protein translation while it is encoding tRNA. This can trigger ribosomal frameshifting.
199:
ternary complex (eIF2-TC). When large numbers of eIF2 are phosphorylated, protein synthesis is inhibited. This occurs under amino acid starvation or after viral infection. However, a small fraction of this initiation factor is naturally phosphorylated. Another regulator is
253:
244:. This method of translation is important in conditions that require the translation of specific mRNAs during cellular stress, when overall translation is reduced. Examples include factors responding to apoptosis and stress-induced responses.
216:
settings including yeast meiosis and ethylene response in plants. In addition, recent work in yeast and humans suggest that evolutionary divergence in cis-regulatory sequences can impact translation regulation. Additionally, RNA
928:
Pataskar, Abhijeet; Champagne, Julien; Nagel, Remco; Kenski, Juliana; Laos, Maarja; Michaux, Justine; Pak, Hui Song; Bleijerveld, Onno B.; Mordente, Kelly; Navarro, Jasmine
Montenegro; Blommaert, Naomi (2022-03-24).
264:. At the end of the initiation step, the mRNA is positioned so that the next codon can be translated during the elongation stage of protein synthesis. The initiator tRNA occupies the P site in the
2346:
256:
The elongation and membrane targeting stages of eukaryotic translation. The ribosome is green and yellow, the tRNAs are dark-blue, and the other proteins involved are light-blue
212:, thus preventing cap-dependent initiation. To oppose the effects of 4EBP, growth factors phosphorylate 4EBP, reducing its affinity for eIF4E and permitting protein synthesis.
180:. It hydrolyzes GTP, and signals for the dissociation of several factors from the small ribosomal subunit, eventually leading to the association of the large subunit (or the
1997:
1821:
914:
332:
Translation is one of the key energy consumers in cells, hence it is strictly regulated. Numerous mechanisms have evolved that control and regulate translation in
104:
is associated with the 40S ribosomal subunit and plays a role in keeping the large (60S) ribosomal subunit from prematurely binding. eIF3 also interacts with the
1408:
2376:
1021:
2386:
2123:
989:
2423:
2419:
1189:
577:
Cenik C, Cenik ES, Byeon GW, Grubert F, Candille SI, Spacek D, Alsallakh B, Tilgner H, Araya CL, Tang H, Ricci E, Snyder MP (November 2015).
152:(43S PIC) accompanied by the protein factors moves along the mRNA chain toward its 3'-end, in a process known as 'scanning', to reach the
145:
of most eukaryotic mRNA molecules. This protein has been implicated in playing a role in circularization of the mRNA during translation.
2798:
2793:
2788:
2783:
2773:
2753:
2748:
2738:
2728:
2708:
2703:
2698:
2688:
2683:
2678:
2673:
2663:
2653:
2648:
2643:
2638:
2415:
133:
is an ATP-dependent RNA helicase that aids the ribosome by resolving certain secondary structures formed along the mRNA transcript. The
2608:
2603:
2593:
1476:
1073:
1053:
1401:
1161:
376:
instead of tryptophan. The resulting peptides are called W>F "substitutants". Such W>F substitutants are abundant in certain
2366:
2361:
2356:
1471:
1466:
1315:
1259:
105:
2181:
1254:
101:
3062:
1342:
1273:
1014:
413:
261:
2778:
2768:
2763:
2758:
2743:
2733:
2723:
2718:
2713:
2693:
2668:
2658:
2633:
2628:
2623:
2618:
2613:
2598:
1547:
1394:
1078:
408:
237:
3072:
1514:
1458:
1386:
361:
3057:
2544:
2484:
2331:
2321:
2311:
2271:
2251:
2226:
2201:
2176:
1293:
418:
301:
1537:
1532:
1527:
1522:
579:"Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans"
3012:
2559:
2554:
2549:
2539:
2534:
2529:
2524:
2519:
2514:
2509:
2504:
2499:
2494:
2489:
2479:
2474:
2469:
2464:
2459:
2454:
2409:
2351:
2341:
2336:
2326:
2316:
2306:
2301:
2296:
2291:
2286:
2281:
2276:
2266:
2261:
2256:
2246:
2241:
2236:
2231:
2221:
2216:
2211:
2206:
2196:
2191:
2186:
2171:
2156:
149:
3067:
3036:
2449:
2444:
2439:
2429:
2404:
2399:
2166:
2161:
2151:
2146:
2141:
2136:
2131:
1337:
1125:
1038:
1007:
677:"Protein synthesis in eukaryotes: the growing biological relevance of cap-independent translation initiation"
380:
types and have been associated with increased IDO1 expression. Functionally, W>F substitutants can impair
3032:
1298:
1115:
1100:
134:
2578:
1303:
1120:
1425:
1421:
1218:
1203:
1083:
908:
305:
38:
228:
may participate in the process of translation initiation, especially for mRNAs with structured 5'UTRs.
994:
57:
2434:
2394:
1429:
1417:
1325:
1223:
1141:
821:
1959:
1151:
628:"Translation initiation on mammalian mRNAs with structured 5'UTRs requires DExH-box protein DHX29"
1286:
1169:
810:"Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling"
477:
17:
124:
is a scaffolding protein that directly associates with both eIF3 and the other two components.
2105:
1922:
1449:
1370:
968:
950:
896:
847:
790:
739:
698:
657:
608:
559:
518:
469:
83:
2979:
2969:
2964:
2954:
2944:
2924:
2909:
2904:
2899:
2884:
2879:
2874:
2859:
2007:
958:
942:
886:
878:
837:
829:
780:
770:
729:
688:
647:
639:
598:
590:
549:
508:
459:
451:
357:
2849:
2829:
2824:
2814:
1504:
1494:
129:
translation to hijack the host machinery in favor of the viral (cap-independent) messages.
1347:
1184:
1030:
289:
281:
236:
The best-studied example of cap-independent translation initiation in eukaryotes uses the
891:
866:
825:
464:
439:
82:
Initiation of translation usually involves the interaction of certain key proteins, the
2067:
1174:
1146:
963:
930:
842:
809:
785:
758:
718:"Halting a cellular production line: responses to ribosomal pausing during translation"
652:
627:
603:
578:
317:
309:
554:
537:
316:
of up to 10% of the stop codons of these genes. Some of these genes encode functional
3051:
3017:
1375:
1062:
373:
34:
693:
676:
481:
1619:
1486:
1249:
1179:
1048:
369:
277:
269:
252:
49:. It consists of four phases: initiation, elongation, termination, and recapping.
775:
3022:
1352:
1281:
352:
can be depleted and thus affect translation efficiency. For instance, activated
324:
can arise. This process has been termed 'functional translational readthrough'.
153:
142:
946:
882:
643:
3027:
1310:
455:
365:
349:
337:
313:
169:
165:
46:
954:
867:"Single-cell quantification of ribosome occupancy in early mouse development"
2114:
833:
333:
285:
273:
157:
972:
931:"Tryptophan depletion results in tryptophan-to-phenylalanine substitutants"
900:
851:
794:
743:
702:
661:
612:
522:
473:
241:
191:
Regulation of protein synthesis is partly influenced by phosphorylation of
91:
87:
865:
Ozadam H, Tonn T, Han CM, Segura A, Hoskins I, Rao S; et al. (2023).
626:
Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV (December 2008).
594:
563:
2029:
2024:
2019:
1330:
1320:
1244:
265:
217:
201:
95:
440:"Translation initiation: variations in the mechanism can be anticipated"
1750:
1066:
734:
717:
513:
496:
381:
321:
161:
42:
538:"Circularization of mRNA by eukaryotic translation initiation factors"
74:
2984:
2974:
2959:
2949:
2939:
2934:
2929:
2919:
2914:
2894:
2889:
2869:
2864:
2041:
2014:
2002:
1875:
1831:
1826:
1809:
1804:
1792:
1787:
1782:
1715:
1666:
1661:
1656:
1651:
1646:
1631:
1626:
1614:
1590:
1585:
759:"Functional Translational Readthrough: A Systems Biology Perspective"
377:
360:
which triggers intracellular tryptophan shortage by upregulating the
353:
2854:
2844:
2839:
2834:
2819:
2564:
2371:
2093:
2088:
2046:
2036:
1992:
1944:
1939:
1882:
1870:
1843:
1838:
1816:
1799:
1777:
1755:
1745:
1740:
1735:
1730:
1725:
1720:
1710:
1705:
1700:
1695:
1641:
1636:
1573:
1499:
1088:
999:
368:
depletion, in-frame protein synthesis continues across tryptophan
251:
225:
221:
209:
205:
138:
130:
125:
121:
117:
113:
109:
86:, with a special tag bound to the 5'-end of an mRNA molecule, the
73:
56:
808:
Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (April 2009).
2078:
2053:
1974:
1969:
1964:
1954:
1949:
1904:
1894:
1865:
1855:
1767:
1685:
1673:
1604:
1578:
1568:
1558:
192:
177:
1390:
1003:
1058:
403:
398:
393:
185:
181:
195:(via the α subunit), which is a part of the eIF2-GTP-Met-tRNA
176:) is brought to the P-site of the small ribosomal subunit by
497:"Internal ribosome entry sites in eukaryotic mRNA molecules"
108:
complex, which consists of three other initiation factors:
308:, but unlike bacterial termination, there is a universal
284:, splicing) in ribosomes before they are exported to the
276:). Eukaryotic mRNA precursors must be processed in the
66:
where the ribosome does not initially bind the 5' cap.
61:
The process of initiation of translation in eukaryotes.
536:
Wells SE, Hillner PE, Vale RD, Sachs AB (July 1998).
288:
for translation. Translation can also be affected by
78:
some of the protein complexes involved in initiation
3005:
2807:
2586:
2577:
2385:
2122:
2113:
2104:
2066:
1983:
1930:
1921:
1891:
1852:
1764:
1682:
1601:
1555:
1546:
1513:
1485:
1457:
1448:
1441:
1363:
1272:
1237:
1211:
1202:
1160:
1134:
1108:
1099:
1037:
320:in their readthrough extension so that new protein
1402:
1015:
8:
913:: CS1 maint: multiple names: authors list (
675:López-Lastra M, Rivas A, Barría MI (2005).
2583:
2119:
2110:
1927:
1552:
1454:
1445:
1409:
1395:
1387:
1208:
1170:Precursor mRNA (pre-mRNA / hnRNA)
1105:
1022:
1008:
1000:
716:Buchan JR, Stansfield I (September 2007).
328:Regulation and modification of translation
172:. The Met-charged initiator tRNA (Met-tRNA
962:
890:
841:
784:
774:
733:
692:
651:
602:
553:
512:
463:
188:) then commences translation elongation.
430:
372:. This is achieved by incorporation of
364:1 (IDO1) enzyme. Surprisingly, despite
204:, which binds to the initiation factor
906:
94:. These proteins bind the small (40S)
1190:Histone acetylation and deacetylation
300:Termination of elongation depends on
178:eukaryotic initiation factor 2 (eIF2)
7:
1255:Ribosome-nascent chain complex (RNC)
444:Cellular and Molecular Life Sciences
304:. The process is similar to that of
98:subunit and hold the mRNA in place.
757:Schueren F, Thoms S (August 2016).
438:Malys N, McCarthy JE (March 2011).
208:and inhibits its interactions with
33:is the biological process by which
25:
495:Hellen CU, Sarnow P (July 2001).
184:subunit). The complete ribosome (
141:complex via eIF4G, and binds the
18:Translation preinitiation complex
137:(PABP) also associates with the
1260:Post-translational modification
694:10.4067/s0716-97602005000200003
168:encoded by the start codon is
1:
555:10.1016/S1097-2765(00)80122-7
414:Eukaryotic elongation factors
262:eukaryotic elongation factors
995:Animations at nobelprize.org
776:10.1371/JOURNAL.PGEN.1006196
409:Eukaryotic initiation factor
238:internal ribosome entry site
362:indoleamine 2,3-dioxygenase
3089:
947:10.1038/s41586-022-04499-2
883:10.1038/s41586-023-06228-9
644:10.1016/j.cell.2008.10.037
419:Eukaryotic release factors
302:eukaryotic release factors
232:Cap-independent initiation
3013:Aminoacyl tRNA synthetase
456:10.1007/s00018-010-0588-z
314:translational readthrough
150:43S preinitiation complex
3037:Kozak consensus sequence
1321:sequestration (P-bodies)
70:Cap-dependent initiation
3033:Shine-Dalgarno sequence
1299:Gene regulatory network
834:10.1126/science.1168978
501:Genes & Development
344:Amino acid substitution
135:poly(A)-binding protein
1304:cis-regulatory element
348:In some cells certain
260:Elongation depends on
257:
90:, as well as with the
79:
62:
31:Eukaryotic translation
595:10.1101/gr.193342.115
306:bacterial termination
255:
77:
60:
3063:Protein biosynthesis
1933:Mitochondrial
1418:Protein biosynthesis
1326:alternative splicing
1316:Post-transcriptional
1142:Transcription factor
990:Animation at wku.edu
156:(typically AUG). In
1250:Transfer RNA (tRNA)
877:(7967): 1057–1064.
826:2009Sci...324..218I
722:Biology of the Cell
681:Biological Research
3073:Eukaryote genetics
2106:Ribosomal Proteins
1364:Influential people
1343:Post-translational
1162:Post-transcription
735:10.1042/BC20070037
514:10.1101/gad.891101
258:
84:initiation factors
80:
63:
3058:Molecular biology
3045:
3044:
3001:
3000:
2997:
2996:
2993:
2992:
2573:
2572:
2062:
2061:
1986:Eukaryotic
1923:Elongation factor
1917:
1916:
1913:
1912:
1450:Initiation factor
1384:
1383:
1268:
1267:
1198:
1197:
1074:Special transfers
941:(7902): 721–727.
290:ribosomal pausing
16:(Redirected from
3080:
2584:
2120:
2111:
1985:
1932:
1928:
1898:
1859:
1771:
1689:
1608:
1562:
1553:
1455:
1446:
1411:
1404:
1397:
1388:
1209:
1106:
1024:
1017:
1010:
1001:
977:
976:
966:
925:
919:
918:
912:
904:
894:
862:
856:
855:
845:
820:(5924): 218–23.
805:
799:
798:
788:
778:
754:
748:
747:
737:
713:
707:
706:
696:
672:
666:
665:
655:
623:
617:
616:
606:
574:
568:
567:
557:
533:
527:
526:
516:
507:(13): 1593–612.
492:
486:
485:
467:
435:
280:(e.g., capping,
21:
3088:
3087:
3083:
3082:
3081:
3079:
3078:
3077:
3068:Gene expression
3048:
3047:
3046:
3041:
2989:
2803:
2569:
2381:
2100:
2058:
1979:
1909:
1892:
1887:
1853:
1848:
1765:
1760:
1683:
1678:
1602:
1597:
1556:
1542:
1509:
1481:
1437:
1415:
1385:
1380:
1359:
1294:Transcriptional
1264:
1233:
1194:
1185:Polyadenylation
1156:
1130:
1095:
1089:Protein→Protein
1040:
1033:
1031:Gene expression
1028:
986:
981:
980:
927:
926:
922:
905:
864:
863:
859:
807:
806:
802:
769:(8): e1006196.
756:
755:
751:
715:
714:
710:
687:(2–3): 121–46.
674:
673:
669:
625:
624:
620:
589:(11): 1610–21.
583:Genome Research
576:
575:
571:
535:
534:
530:
494:
493:
489:
450:(6): 991–1003.
437:
436:
432:
427:
390:
346:
330:
318:protein domains
298:
282:polyadenylation
250:
234:
198:
175:
72:
55:
28:
27:RNS translation
23:
22:
15:
12:
11:
5:
3086:
3084:
3076:
3075:
3070:
3065:
3060:
3050:
3049:
3043:
3042:
3040:
3039:
3030:
3025:
3020:
3015:
3009:
3007:
3006:Other concepts
3003:
3002:
2999:
2998:
2995:
2994:
2991:
2990:
2988:
2987:
2982:
2977:
2972:
2967:
2962:
2957:
2952:
2947:
2942:
2937:
2932:
2927:
2922:
2917:
2912:
2907:
2902:
2897:
2892:
2887:
2882:
2877:
2872:
2867:
2862:
2857:
2852:
2847:
2842:
2837:
2832:
2827:
2822:
2817:
2811:
2809:
2805:
2804:
2802:
2801:
2796:
2791:
2786:
2781:
2776:
2771:
2766:
2761:
2756:
2751:
2746:
2741:
2736:
2731:
2726:
2721:
2716:
2711:
2706:
2701:
2696:
2691:
2686:
2681:
2676:
2671:
2666:
2661:
2656:
2651:
2646:
2641:
2636:
2631:
2626:
2621:
2616:
2611:
2606:
2601:
2596:
2590:
2588:
2581:
2575:
2574:
2571:
2570:
2568:
2567:
2562:
2557:
2552:
2547:
2542:
2537:
2532:
2527:
2522:
2517:
2512:
2507:
2502:
2497:
2492:
2487:
2482:
2477:
2472:
2467:
2462:
2457:
2452:
2447:
2442:
2437:
2432:
2427:
2412:
2407:
2402:
2397:
2391:
2389:
2383:
2382:
2380:
2379:
2374:
2369:
2364:
2359:
2354:
2349:
2344:
2339:
2334:
2329:
2324:
2319:
2314:
2309:
2304:
2299:
2294:
2289:
2284:
2279:
2274:
2269:
2264:
2259:
2254:
2249:
2244:
2239:
2234:
2229:
2224:
2219:
2214:
2209:
2204:
2199:
2194:
2189:
2184:
2179:
2174:
2169:
2164:
2159:
2154:
2149:
2144:
2139:
2134:
2128:
2126:
2117:
2108:
2102:
2101:
2099:
2098:
2097:
2096:
2091:
2083:
2082:
2081:
2072:
2070:
2068:Release factor
2064:
2063:
2060:
2059:
2057:
2056:
2051:
2050:
2049:
2044:
2039:
2034:
2033:
2032:
2027:
2022:
2012:
2011:
2010:
2005:
1989:
1987:
1981:
1980:
1978:
1977:
1972:
1967:
1962:
1957:
1952:
1947:
1942:
1936:
1934:
1925:
1919:
1918:
1915:
1914:
1911:
1910:
1908:
1907:
1901:
1899:
1889:
1888:
1886:
1885:
1880:
1879:
1878:
1868:
1862:
1860:
1850:
1849:
1847:
1846:
1841:
1836:
1835:
1834:
1829:
1824:
1814:
1813:
1812:
1807:
1797:
1796:
1795:
1790:
1785:
1774:
1772:
1762:
1761:
1759:
1758:
1753:
1748:
1743:
1738:
1733:
1728:
1723:
1718:
1713:
1708:
1703:
1698:
1692:
1690:
1680:
1679:
1677:
1676:
1671:
1670:
1669:
1664:
1659:
1654:
1649:
1639:
1634:
1629:
1624:
1623:
1622:
1611:
1609:
1599:
1598:
1596:
1595:
1594:
1593:
1583:
1582:
1581:
1576:
1565:
1563:
1550:
1544:
1543:
1541:
1540:
1535:
1530:
1525:
1519:
1517:
1511:
1510:
1508:
1507:
1502:
1497:
1491:
1489:
1483:
1482:
1480:
1479:
1474:
1469:
1463:
1461:
1452:
1443:
1439:
1438:
1416:
1414:
1413:
1406:
1399:
1391:
1382:
1381:
1379:
1378:
1373:
1371:François Jacob
1367:
1365:
1361:
1360:
1358:
1357:
1356:
1355:
1350:
1340:
1335:
1334:
1333:
1328:
1323:
1313:
1308:
1307:
1306:
1301:
1291:
1290:
1289:
1278:
1276:
1270:
1269:
1266:
1265:
1263:
1262:
1257:
1252:
1247:
1241:
1239:
1235:
1234:
1232:
1231:
1226:
1221:
1215:
1213:
1206:
1200:
1199:
1196:
1195:
1193:
1192:
1187:
1182:
1177:
1172:
1166:
1164:
1158:
1157:
1155:
1154:
1149:
1147:RNA polymerase
1144:
1138:
1136:
1132:
1131:
1129:
1128:
1123:
1118:
1112:
1110:
1103:
1097:
1096:
1094:
1093:
1092:
1091:
1086:
1081:
1071:
1070:
1069:
1051:
1045:
1043:
1035:
1034:
1029:
1027:
1026:
1019:
1012:
1004:
998:
997:
992:
985:
984:External links
982:
979:
978:
920:
857:
800:
749:
708:
667:
638:(7): 1237–50.
618:
569:
542:Molecular Cell
528:
487:
429:
428:
426:
423:
422:
421:
416:
411:
406:
401:
396:
389:
386:
345:
342:
329:
326:
310:release factor
297:
294:
249:
246:
233:
230:
196:
173:
71:
68:
54:
51:
35:messenger RNA
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
3085:
3074:
3071:
3069:
3066:
3064:
3061:
3059:
3056:
3055:
3053:
3038:
3034:
3031:
3029:
3026:
3024:
3021:
3019:
3018:Reading frame
3016:
3014:
3011:
3010:
3008:
3004:
2986:
2983:
2981:
2978:
2976:
2973:
2971:
2968:
2966:
2963:
2961:
2958:
2956:
2953:
2951:
2948:
2946:
2943:
2941:
2938:
2936:
2933:
2931:
2928:
2926:
2923:
2921:
2918:
2916:
2913:
2911:
2908:
2906:
2903:
2901:
2898:
2896:
2893:
2891:
2888:
2886:
2883:
2881:
2878:
2876:
2873:
2871:
2868:
2866:
2863:
2861:
2858:
2856:
2853:
2851:
2848:
2846:
2843:
2841:
2838:
2836:
2833:
2831:
2828:
2826:
2823:
2821:
2818:
2816:
2813:
2812:
2810:
2806:
2800:
2797:
2795:
2792:
2790:
2787:
2785:
2782:
2780:
2777:
2775:
2772:
2770:
2767:
2765:
2762:
2760:
2757:
2755:
2752:
2750:
2747:
2745:
2742:
2740:
2737:
2735:
2732:
2730:
2727:
2725:
2722:
2720:
2717:
2715:
2712:
2710:
2707:
2705:
2702:
2700:
2697:
2695:
2692:
2690:
2687:
2685:
2682:
2680:
2677:
2675:
2672:
2670:
2667:
2665:
2662:
2660:
2657:
2655:
2652:
2650:
2647:
2645:
2642:
2640:
2637:
2635:
2632:
2630:
2627:
2625:
2622:
2620:
2617:
2615:
2612:
2610:
2607:
2605:
2602:
2600:
2597:
2595:
2592:
2591:
2589:
2585:
2582:
2580:
2579:Mitochondrial
2576:
2566:
2563:
2561:
2558:
2556:
2553:
2551:
2548:
2546:
2543:
2541:
2538:
2536:
2533:
2531:
2528:
2526:
2523:
2521:
2518:
2516:
2513:
2511:
2508:
2506:
2503:
2501:
2498:
2496:
2493:
2491:
2488:
2486:
2483:
2481:
2478:
2476:
2473:
2471:
2468:
2466:
2463:
2461:
2458:
2456:
2453:
2451:
2448:
2446:
2443:
2441:
2438:
2436:
2433:
2431:
2428:
2425:
2421:
2417:
2413:
2411:
2408:
2406:
2403:
2401:
2398:
2396:
2393:
2392:
2390:
2388:
2384:
2378:
2375:
2373:
2370:
2368:
2365:
2363:
2360:
2358:
2355:
2353:
2350:
2348:
2345:
2343:
2340:
2338:
2335:
2333:
2330:
2328:
2325:
2323:
2320:
2318:
2315:
2313:
2310:
2308:
2305:
2303:
2300:
2298:
2295:
2293:
2290:
2288:
2285:
2283:
2280:
2278:
2275:
2273:
2270:
2268:
2265:
2263:
2260:
2258:
2255:
2253:
2250:
2248:
2245:
2243:
2240:
2238:
2235:
2233:
2230:
2228:
2225:
2223:
2220:
2218:
2215:
2213:
2210:
2208:
2205:
2203:
2200:
2198:
2195:
2193:
2190:
2188:
2185:
2183:
2180:
2178:
2175:
2173:
2170:
2168:
2165:
2163:
2160:
2158:
2155:
2153:
2150:
2148:
2145:
2143:
2140:
2138:
2135:
2133:
2130:
2129:
2127:
2125:
2121:
2118:
2116:
2112:
2109:
2107:
2103:
2095:
2092:
2090:
2087:
2086:
2084:
2080:
2077:
2076:
2074:
2073:
2071:
2069:
2065:
2055:
2052:
2048:
2045:
2043:
2040:
2038:
2035:
2031:
2028:
2026:
2023:
2021:
2018:
2017:
2016:
2013:
2009:
2006:
2004:
2001:
2000:
1999:
1996:
1995:
1994:
1991:
1990:
1988:
1982:
1976:
1973:
1971:
1968:
1966:
1963:
1961:
1958:
1956:
1953:
1951:
1948:
1946:
1943:
1941:
1938:
1937:
1935:
1929:
1926:
1924:
1920:
1906:
1903:
1902:
1900:
1897:
1896:
1890:
1884:
1881:
1877:
1874:
1873:
1872:
1869:
1867:
1864:
1863:
1861:
1858:
1857:
1851:
1845:
1842:
1840:
1837:
1833:
1830:
1828:
1825:
1823:
1820:
1819:
1818:
1815:
1811:
1808:
1806:
1803:
1802:
1801:
1798:
1794:
1791:
1789:
1786:
1784:
1781:
1780:
1779:
1776:
1775:
1773:
1770:
1769:
1763:
1757:
1754:
1752:
1749:
1747:
1744:
1742:
1739:
1737:
1734:
1732:
1729:
1727:
1724:
1722:
1719:
1717:
1714:
1712:
1709:
1707:
1704:
1702:
1699:
1697:
1694:
1693:
1691:
1688:
1687:
1681:
1675:
1672:
1668:
1665:
1663:
1660:
1658:
1655:
1653:
1650:
1648:
1645:
1644:
1643:
1640:
1638:
1635:
1633:
1630:
1628:
1625:
1621:
1618:
1617:
1616:
1613:
1612:
1610:
1607:
1606:
1600:
1592:
1589:
1588:
1587:
1584:
1580:
1577:
1575:
1572:
1571:
1570:
1567:
1566:
1564:
1561:
1560:
1554:
1551:
1549:
1545:
1539:
1536:
1534:
1531:
1529:
1526:
1524:
1521:
1520:
1518:
1516:
1512:
1506:
1503:
1501:
1498:
1496:
1493:
1492:
1490:
1488:
1487:Mitochondrial
1484:
1478:
1475:
1473:
1470:
1468:
1465:
1464:
1462:
1460:
1456:
1453:
1451:
1447:
1444:
1440:
1435:
1431:
1427:
1423:
1419:
1412:
1407:
1405:
1400:
1398:
1393:
1392:
1389:
1377:
1376:Jacques Monod
1374:
1372:
1369:
1368:
1366:
1362:
1354:
1351:
1349:
1346:
1345:
1344:
1341:
1339:
1338:Translational
1336:
1332:
1329:
1327:
1324:
1322:
1319:
1318:
1317:
1314:
1312:
1309:
1305:
1302:
1300:
1297:
1296:
1295:
1292:
1288:
1285:
1284:
1283:
1280:
1279:
1277:
1275:
1271:
1261:
1258:
1256:
1253:
1251:
1248:
1246:
1243:
1242:
1240:
1236:
1230:
1227:
1225:
1222:
1220:
1217:
1216:
1214:
1210:
1207:
1205:
1201:
1191:
1188:
1186:
1183:
1181:
1178:
1176:
1173:
1171:
1168:
1167:
1165:
1163:
1159:
1153:
1150:
1148:
1145:
1143:
1140:
1139:
1137:
1133:
1127:
1124:
1122:
1119:
1117:
1114:
1113:
1111:
1107:
1104:
1102:
1101:Transcription
1098:
1090:
1087:
1085:
1082:
1080:
1077:
1076:
1075:
1072:
1068:
1064:
1060:
1057:
1056:
1055:
1054:Central dogma
1052:
1050:
1047:
1046:
1044:
1042:
1036:
1032:
1025:
1020:
1018:
1013:
1011:
1006:
1005:
1002:
996:
993:
991:
988:
987:
983:
974:
970:
965:
960:
956:
952:
948:
944:
940:
936:
932:
924:
921:
916:
910:
902:
898:
893:
888:
884:
880:
876:
872:
868:
861:
858:
853:
849:
844:
839:
835:
831:
827:
823:
819:
815:
811:
804:
801:
796:
792:
787:
782:
777:
772:
768:
764:
763:PLOS Genetics
760:
753:
750:
745:
741:
736:
731:
728:(9): 475–87.
727:
723:
719:
712:
709:
704:
700:
695:
690:
686:
682:
678:
671:
668:
663:
659:
654:
649:
645:
641:
637:
633:
629:
622:
619:
614:
610:
605:
600:
596:
592:
588:
584:
580:
573:
570:
565:
561:
556:
551:
548:(1): 135–40.
547:
543:
539:
532:
529:
524:
520:
515:
510:
506:
502:
498:
491:
488:
483:
479:
475:
471:
466:
461:
457:
453:
449:
445:
441:
434:
431:
424:
420:
417:
415:
412:
410:
407:
405:
402:
400:
397:
395:
392:
391:
387:
385:
383:
379:
375:
374:phenylalanine
371:
367:
363:
359:
355:
351:
343:
341:
339:
335:
327:
325:
323:
319:
315:
311:
307:
303:
295:
293:
291:
287:
283:
279:
275:
271:
267:
263:
254:
247:
245:
243:
239:
231:
229:
227:
223:
219:
213:
211:
207:
203:
194:
189:
187:
183:
179:
171:
167:
163:
159:
155:
151:
146:
144:
140:
136:
132:
127:
123:
119:
115:
111:
107:
103:
99:
97:
93:
89:
85:
76:
69:
67:
59:
52:
50:
48:
44:
40:
36:
32:
19:
2085:Class 2/RF3
1893:
1854:
1766:
1684:
1603:
1557:
1433:
1353:irreversible
1238:Key elements
1228:
1135:Key elements
1049:Genetic code
1039:Introduction
938:
934:
923:
909:cite journal
874:
870:
860:
817:
813:
803:
766:
762:
752:
725:
721:
711:
684:
680:
670:
635:
631:
621:
586:
582:
572:
545:
541:
531:
504:
500:
490:
447:
443:
433:
358:interferon-γ
347:
331:
299:
259:
235:
214:
190:
147:
100:
81:
64:
30:
29:
3023:Start codon
2808:28S subunit
2587:39S subunit
2387:40S subunit
2124:60S subunit
2115:Cytoplasmic
1579:SUI1 family
1422:translation
1204:Translation
1041:to genetics
350:amino acids
338:prokaryotes
336:as well as
296:Termination
154:start codon
143:poly-A tail
3052:Categories
3028:Stop codon
2372:RRP15-like
2182:RPL10-like
1931:Bacterial/
1548:Eukaryotic
1434:eukaryotic
1348:reversible
1311:lac operon
1287:imprinting
1282:Epigenetic
1274:Regulation
1229:Eukaryotic
1175:5' capping
1126:Eukaryotic
425:References
384:activity.
366:tryptophan
334:eukaryotes
248:Elongation
170:methionine
166:amino acid
158:eukaryotes
53:Initiation
47:eukaryotes
39:translated
1984:Archaeal/
1459:Bacterial
1426:bacterial
1219:Bacterial
1116:Bacterial
955:0028-0836
286:cytoplasm
274:cytoplasm
226:Ded1/DDX3
218:helicases
96:ribosomal
2075:Class 1
1515:Archaeal
1442:Proteins
1430:archaeal
1331:microRNA
1245:Ribosome
1224:Archaeal
1180:Splicing
1152:Promoter
1121:Archaeal
1065: →
1061: →
973:35264796
901:37344592
892:10307641
852:19213877
795:27490485
744:17696878
703:16238092
662:19109895
613:26297486
523:11445534
482:31720000
474:21076851
465:11115079
388:See also
356:secrete
322:isoforms
266:ribosome
220:such as
43:proteins
2377:RSL24D1
2054:a/eEF-2
1993:a/eEF-1
1084:RNA→DNA
1079:RNA→RNA
1067:Protein
964:8942854
843:2746483
822:Bibcode
814:Science
786:4973966
653:2948571
604:4617958
564:9702200
382:protein
354:T cells
278:nucleus
270:nucleus
162:archaea
2545:RPS27A
2485:RPS15A
2424:RPS4Y2
2420:RPS4Y1
2414:RPS4 (
2332:RPL37A
2322:RPL36A
2312:RPL35A
2272:RPL27A
2252:RPL23A
2227:RPL18A
2202:RPL13A
2177:RPL10A
1632:γ
1627:β
1620:kinase
1615:α
971:
961:
953:
935:Nature
899:
889:
871:Nature
850:
840:
793:
783:
742:
701:
660:
650:
611:
601:
562:
521:
480:
472:
462:
378:cancer
370:codons
242:5' UTR
164:, the
116:, and
92:5' UTR
88:5' cap
2815:MRPS1
2594:MRPL1
2565:RACK1
2560:RPS30
2555:RPS29
2550:RPS28
2540:RPS27
2535:RPS26
2530:RPS25
2525:RPS24
2520:RPS23
2515:RPS21
2510:RPS20
2505:RPS19
2500:RPS18
2495:RPS17
2490:RPS16
2480:RPS15
2475:RPS14
2470:RPS13
2465:RPS12
2460:RPS11
2455:RPS10
2416:RPS4X
2410:RPS3A
2367:RPLP2
2362:RPLP1
2357:RPLP0
2352:RPL41
2347:RPL40
2342:RPL39
2337:RPL38
2327:RPL37
2317:RPL36
2307:RPL35
2302:RPL34
2297:RPL32
2292:RPL31
2287:RPL30
2282:RPL29
2277:RPL28
2267:RPL27
2262:RPL26
2257:RPL24
2247:RPL23
2242:RPL22
2237:RPL21
2232:RPL19
2222:RPL18
2217:RPL17
2212:RPL15
2207:RPL14
2197:RPL13
2192:RPL12
2187:RPL11
2172:RPL10
2157:RPL7A
2094:GSPT2
2089:GSPT1
1945:EF-Ts
1940:EF-Tu
1871:EIF5A
1674:eIF2D
1642:eIF2B
1637:eIF2A
1586:eIF1A
1505:MTIF3
1500:MTIF2
1495:MTIF1
1212:Types
1109:Types
478:S2CID
222:DHX29
210:eIF4G
206:eIF4E
148:This
139:eIF4F
131:eIF4A
126:eIF4E
122:eIF4G
118:eIF4G
114:eIF4E
110:eIF4A
106:eIF4F
41:into
2450:RPS9
2445:RPS8
2440:RPS7
2435:RPS6
2430:RPS5
2405:RPS3
2400:RPS2
2395:RPSA
2167:RPL9
2162:RPL8
2152:RPL7
2147:RPL6
2142:RPL5
2137:RPL4
2132:RPL3
2079:eRF1
1975:GFM2
1970:GFM1
1965:TSFM
1960:EF-P
1955:EF-4
1950:EF-G
1905:EIF6
1895:eIF6
1866:EIF5
1856:eIF5
1768:eIF4
1686:eIF3
1605:eIF2
1569:eIF1
1559:eIF1
1538:aIF6
1533:aIF5
1528:aIF2
1523:aIF1
969:PMID
951:ISSN
915:link
897:PMID
848:PMID
791:PMID
740:PMID
699:PMID
658:PMID
632:Cell
609:PMID
560:PMID
519:PMID
470:PMID
272:and
224:and
202:4EBP
193:eIF2
160:and
102:eIF3
1477:IF3
1472:IF2
1467:IF1
1063:RNA
1059:DNA
959:PMC
943:doi
939:603
887:PMC
879:doi
875:618
838:PMC
830:doi
818:324
781:PMC
771:doi
730:doi
689:doi
648:PMC
640:doi
636:135
599:PMC
591:doi
550:doi
509:doi
460:PMC
452:doi
404:80S
399:60S
394:40S
186:80S
182:60S
45:in
37:is
3054::
2985:35
2980:34
2975:33
2970:32
2965:31
2960:30
2955:29
2950:28
2945:27
2940:26
2935:25
2930:24
2925:23
2920:22
2915:21
2910:20
2905:19
2900:18
2895:17
2890:16
2885:15
2880:14
2875:13
2870:12
2865:11
2860:10
2799:42
2794:41
2789:40
2784:39
2779:38
2774:37
2769:36
2764:35
2759:34
2754:33
2749:32
2744:31
2739:30
2734:29
2729:28
2724:27
2719:26
2714:25
2709:24
2704:23
2699:22
2694:21
2689:20
2684:19
2679:18
2674:17
2669:16
2664:15
2659:14
2654:13
2649:12
2644:11
2639:10
2422:,
2418:,
2030:P3
2025:P2
2020:P1
1998:A1
1883:5B
1800:E1
1432:,
1428:,
1420::
967:.
957:.
949:.
937:.
933:.
911:}}
907:{{
895:.
885:.
873:.
869:.
846:.
836:.
828:.
816:.
812:.
789:.
779:.
767:12
765:.
761:.
738:.
726:99
724:.
720:.
697:.
685:38
683:.
679:.
656:.
646:.
634:.
630:.
607:.
597:.
587:25
585:.
581:.
558:.
544:.
540:.
517:.
505:15
503:.
499:.
476:.
468:.
458:.
448:68
446:.
442:.
120:.
112:,
3035:/
2855:9
2850:8
2845:7
2840:6
2835:5
2830:4
2825:3
2820:2
2634:9
2629:8
2624:7
2619:6
2614:5
2609:4
2604:3
2599:2
2426:)
2047:G
2042:E
2037:D
2015:B
2008:3
2003:2
1876:2
1844:H
1839:B
1832:3
1827:2
1822:1
1817:G
1810:3
1805:2
1793:3
1788:2
1783:1
1778:A
1756:M
1751:L
1746:K
1741:J
1736:I
1731:H
1726:G
1721:F
1716:E
1711:D
1706:C
1701:B
1696:A
1667:5
1662:4
1657:3
1652:2
1647:1
1591:Y
1574:B
1436:)
1424:(
1410:e
1403:t
1396:v
1023:e
1016:t
1009:v
975:.
945::
917:)
903:.
881::
854:.
832::
824::
797:.
773::
746:.
732::
705:.
691::
664:.
642::
615:.
593::
566:.
552::
546:2
525:.
511::
484:.
454::
197:i
174:i
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.