33:
107:
182:
which are commonly used to differentiate the CDRs from each other. The structural relationship between different length CDRS is based on length-independent components, such as their sequence, and can further characterize CDRs. The loops, or three-dimensional structures of the non-H3 CDRs (all CDRs but H3) of antibodies have been clustered and classified by
Chothia et al. and more recently by North et al.
139:), there are six CDRs for each antigen receptor that can collectively come into contact with the antigen. A single antibody molecule has two antigen receptors and therefore contains twelve CDRs total. There are three CDR loops per variable domain in antibodies. Sixty CDRs can be found on a pentameric
181:
of an antibody is important to analyze and design new antibodies. The structure and sequence of all six CDRs combined will determine the binding activity of the antigen receptor on an antibody or T-cell
Receptor. CDRs have been separated into canonical classes based on their varying loop lengths,
168:
Other factors contribute to the antibody-antigen interaction, including amino acid residues. Residues located in particular positions of a CDR loop are used to classify canonical structures. Uncharged-polar residues, especially Serine and
Tyrosine, are found in CDRs at a high concentration ratio.
97:
Antibody-antigen interactions are highly specific and those that have high affinity will interact with increased bond strength and trigger downstream immune responses. The strength of the bond between the epitope of the antigen and the paratope of the antibody will determine the affinity of the
165:. This rearrangement of the V-region is where the CDR-L3 and CDR-H3 are encoded and diversified, whereas the other four CDRs are generated in the germ-line. The diversification of the CDR-H3 will ultimately give antibodies their specificity, and ability to recognize antigens
173:
between the antigen and the antibody. Hydrogen bond interactions will induce the enzymatic activity of an enzyme; therefore, the more hydrogen bonds that are present at the antibody-antigen binding site will result in a stronger, more stable binding structure.
130:
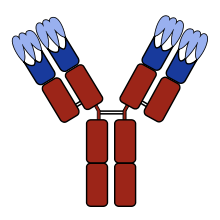
of an antigen receptor. Three can be found on the Light-chain, named L1 through L3, and three on the Heavy-chain, named H1 through H3. Since the antigen receptors are typically composed of two variable domains (on two different polypeptide chains,
160:
of a polypeptide chain, and CDR3 includes some of V, all of diversity (D, heavy chains only) and joining (J) regions. CDR3 is the most variable. The V region sequence undergoes rearrangement during B-cell development, called
81:
respectively. CDRs are where these molecules bind to their specific antigen and their structure/sequence determines the binding activity of the respective antibody. A set of CDRs constitutes a
223:
Osajima T, Suzuki M, Neya S, Hoshino T (September 2014). "Computational and statistical study on the molecular interaction between antigen and antibody".
143:
molecule, which is composed of five antibodies and has increased avidity as a result of the collective affinity of all antigen-binding sites combined.
186:
is a computational method to build tertiary structures from amino-acid sequences. The so-called H3-rules are empirical rules to build models of CDR3.
146:
Since most sequence variation associated with immunoglobulins and T cell receptors are found in the CDRs, these regions are sometimes referred to as
85:, or the antigen-binding site. As the most variable parts of the molecules, CDRs are crucial to the diversity of antigen specificities generated by
351:
326:
260:"Antibody complementarity-determining regions (CDRs) can display differential antimicrobial, antiviral and antitumor activities"
431:
Al-Lazikani B, Lesk AM, Chothia C (November 1997). "Standard conformations for the canonical structures of immunoglobulins".
258:
Polonelli L, Pontón J, Elguezabal N, Moragues MD, Casoli C, Pilotti E, et al. (June 2008). El-Shemy HA (ed.).
136:
132:
41:
571:
567:
162:
32:
592:
528:
385:
271:
200:
148:
374:"Antibody complementarity-determining regions (CDRs): a bridge between adaptive and innate immunity"
372:
Gabrielli E, Pericolini E, Cenci E, Ortelli F, Magliani W, Ciociola T, et al. (December 2009).
602:
597:
123:
17:
178:
546:
497:
448:
413:
347:
322:
299:
240:
183:
46:
536:
487:
479:
440:
403:
393:
289:
279:
232:
195:
127:
115:
70:
66:
532:
389:
275:
492:
467:
408:
373:
294:
259:
541:
516:
586:
170:
118:
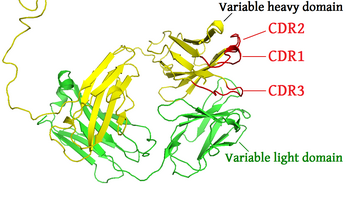
shown in blue, and the CDRs (which are part of the variable domains) in light blue.
106:
398:
284:
86:
236:
122:
There are three CDRs (CDR1, CDR2 and CDR3), arranged non-consecutively, on the
483:
37:
550:
501:
444:
417:
303:
244:
452:
577:
111:
82:
78:
74:
50:
27:
Part of the variable chains in immunoglobulins and T cell receptors
105:
31:
40:) of an antibody. The complementarity-determining regions of the
578:
PyIgClassify -- server for classification of CDR conformations
140:
517:"H3-rules: identification of CDR-H3 structures in antibodies"
169:
These residues significantly contribute to the direct
468:"A new clustering of antibody CDR loop conformations"
65:) are polypeptide segments of the variable chains in
346:(6th ed.). Lippincott Williams & Wilkins.
466:North B, Lehmann A, Dunbrack RL (February 2011).
156:, CDR1 and CDR2 are found in the variable (V)
225:Journal of Molecular Graphics & Modelling
8:
515:Shirai H, Kidera A, Nakamura H (July 1999).
570:at the U.S. National Library of Medicine
540:
491:
407:
397:
293:
283:
321:(5th ed.). Saunders, Philadelphia.
212:
7:
367:
365:
363:
218:
216:
568:Complementarity+determining+regions
59:Complementarity-determining regions
18:Complementarity-determining regions
25:
319:Cellular and Molecular Immunology
317:Abbas AK, Lichtman AH (2003).
1:
542:10.1016/S0014-5793(99)00821-2
472:Journal of Molecular Biology
433:Journal of Molecular Biology
399:10.1371/journal.pone.0008187
285:10.1371/journal.pone.0002371
619:
237:10.1016/j.jmgm.2014.07.005
484:10.1016/j.jmb.2010.10.030
572:Medical Subject Headings
445:10.1006/jmbi.1997.1354
344:Fundamental Immunology
152:. Within the variable
119:
102:Location and structure
55:
163:somatic recombination
149:hypervariable regions
109:
35:
201:Hypervariable region
533:1999FEBSL.455..188S
390:2009PLoSO...4.8187G
276:2008PLoSO...3.2371P
124:amino acid sequence
179:tertiary structure
120:
56:
44:are shown in red (
36:The "upper" part (
353:978-0-7817-6519-0
184:Homology modeling
69:(antibodies) and
16:(Redirected from
610:
555:
554:
544:
527:(1–2): 188–197.
512:
506:
505:
495:
463:
457:
456:
428:
422:
421:
411:
401:
369:
358:
357:
342:Paul WE (2008).
339:
333:
332:
314:
308:
307:
297:
287:
255:
249:
248:
220:
196:Framework region
116:variable domains
93:Binding Affinity
71:T cell receptors
53:
21:
618:
617:
613:
612:
611:
609:
608:
607:
583:
582:
564:
559:
558:
514:
513:
509:
465:
464:
460:
430:
429:
425:
371:
370:
361:
354:
341:
340:
336:
329:
316:
315:
311:
257:
256:
252:
222:
221:
214:
209:
192:
128:variable domain
104:
98:interaction.
95:
73:, generated by
67:immunoglobulins
45:
28:
23:
22:
15:
12:
11:
5:
616:
614:
606:
605:
600:
595:
585:
584:
581:
580:
575:
563:
562:External links
560:
557:
556:
507:
478:(2): 228–256.
458:
439:(4): 927–948.
423:
359:
352:
334:
327:
309:
250:
211:
210:
208:
205:
204:
203:
198:
191:
188:
171:hydrogen bonds
103:
100:
94:
91:
26:
24:
14:
13:
10:
9:
6:
4:
3:
2:
615:
604:
601:
599:
596:
594:
591:
590:
588:
579:
576:
573:
569:
566:
565:
561:
552:
548:
543:
538:
534:
530:
526:
522:
518:
511:
508:
503:
499:
494:
489:
485:
481:
477:
473:
469:
462:
459:
454:
450:
446:
442:
438:
434:
427:
424:
419:
415:
410:
405:
400:
395:
391:
387:
384:(12): e8187.
383:
379:
375:
368:
366:
364:
360:
355:
349:
345:
338:
335:
330:
328:0-7216-0008-5
324:
320:
313:
310:
305:
301:
296:
291:
286:
281:
277:
273:
269:
265:
261:
254:
251:
246:
242:
238:
234:
230:
226:
219:
217:
213:
206:
202:
199:
197:
194:
193:
189:
187:
185:
180:
175:
172:
166:
164:
159:
155:
151:
150:
144:
142:
138:
134:
129:
125:
117:
113:
110:Sketch of an
108:
101:
99:
92:
90:
88:
84:
80:
76:
72:
68:
64:
60:
52:
48:
43:
39:
34:
30:
19:
524:
521:FEBS Letters
520:
510:
475:
471:
461:
436:
432:
426:
381:
377:
343:
337:
318:
312:
270:(6): e2371.
267:
263:
253:
228:
224:
176:
167:
157:
153:
147:
145:
121:
96:
62:
58:
57:
29:
593:Amino acids
231:: 128–139.
137:light chain
87:lymphocytes
42:heavy chain
603:Immunology
598:Antibodies
587:Categories
207:References
38:Fab region
114:with the
54:).
551:10428499
502:21035459
418:19997599
378:PLOS ONE
304:18545659
264:PLOS ONE
245:25123651
190:See also
112:antibody
83:paratope
529:Bibcode
493:3065967
453:9367782
409:2781551
386:Bibcode
295:2396520
272:Bibcode
79:T-cells
75:B-cells
574:(MeSH)
549:
500:
490:
451:
416:
406:
350:
325:
302:
292:
243:
158:region
154:domain
133:heavy
126:of a
547:PMID
498:PMID
449:PMID
414:PMID
348:ISBN
323:ISBN
300:PMID
241:PMID
177:The
135:and
77:and
63:CDRs
51:1IGT
537:doi
525:455
488:PMC
480:doi
476:406
441:doi
437:273
404:PMC
394:doi
290:PMC
280:doi
233:doi
141:IgM
47:PDB
589::
545:.
535:.
523:.
519:.
496:.
486:.
474:.
470:.
447:.
435:.
412:.
402:.
392:.
380:.
376:.
362:^
298:.
288:.
278:.
266:.
262:.
239:.
229:53
227:.
215:^
89:.
49::
553:.
539::
531::
504:.
482::
455:.
443::
420:.
396::
388::
382:4
356:.
331:.
306:.
282::
274::
268:3
247:.
235::
61:(
20:)
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.