17:
103:
or directly activate phagocytic receptors. Calreticulin, annexin A1, histones, pentraxin-3 and DNA may be released by (and onto the surface of) dying cells to encourage phagocytes to eat these cells, thereby acting as self-opsonins. Eat-me signals, or the opsonins that bind them, are recognised by
74:
The most well characterised eat-me signal is the phospholipid phosphatidylserine. Healthy cells do not expose phosphatidylserine on their surface, whereas dead, dying, infected, injured and some activated cells expose phosphatidylserine on their surface in order to induce phagocytes to phagocytose
437:
432:
92:
287:
259:
36:
104:
phagocytic receptors on phagocytes, inducing engulfment of the cell exposing the eat-me signal.
408:
390:
351:
333:
279:
235:
217:
171:
398:
382:
341:
323:
271:
225:
209:
161:
153:
113:
371:"Putting the brakes on phagocytosis: "don't-eat-me" signaling in physiology and disease"
403:
370:
346:
311:
230:
197:
166:
141:
426:
291:
88:
76:
56:
40:
32:
157:
142:"Beginnings of a Good Apoptotic Meal: The Find-Me and Eat-Me Signaling Pathways"
84:
328:
310:
Cockram, Tom O. J.; Dundee, Jacob M.; Popescu, Alma S.; Brown, Guy C. (2021).
275:
198:"Engulfment signals and the phagocytic machinery for apoptotic cell clearance"
80:
68:
60:
394:
337:
221:
91:, but removal of these residues reveals galactose residues (and subsequently
20:
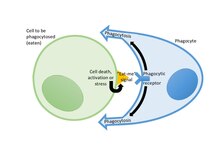
Cells release eat-signals onto their surface to induce phagocytes to eat them
386:
48:
28:
412:
355:
283:
239:
175:
16:
213:
100:
96:
64:
83:
on the surface of our cells have short sugar chains that terminate in
44:
15:
312:"The Phagocytic Code Regulating Phagocytosis of Mammalian Cells"
52:
35:(eat) that cell. Currently known eat-me signals include:
27:
are molecules exposed on the surface of a cell to induce
369:Kelley, Shannon M; Ravichandran, Kodi S (2021).
258:Nagata, Shigekazu; Segawa, Katsumori (2021).
8:
260:"Sensing and clearance of apoptotic cells"
402:
345:
327:
229:
165:
125:
196:Park, Seung-Yoon; Kim, In-San (2017).
202:Experimental & Molecular Medicine
7:
305:
303:
301:
253:
251:
249:
191:
189:
187:
185:
135:
133:
131:
129:
14:
140:Ravichandran, Kodi S. (2011).
1:
264:Current Opinion in Immunology
158:10.1016/j.immuni.2011.09.004
454:
329:10.3389/fimmu.2021.629979
276:10.1016/j.coi.2020.07.007
99:residues) that can bind
87:residues, which inhibit
387:10.15252/embr.202152564
316:Frontiers in Immunology
21:
53:deoxyribonucleic acid
19:
438:Cellular senescence
214:10.1038/emm.2017.52
93:N-acetylglucosamine
47:residues (such as
37:phosphatidylserine
22:
445:
417:
416:
406:
366:
360:
359:
349:
331:
307:
296:
295:
255:
244:
243:
233:
193:
180:
179:
169:
137:
453:
452:
448:
447:
446:
444:
443:
442:
423:
422:
421:
420:
368:
367:
363:
309:
308:
299:
257:
256:
247:
195:
194:
183:
139:
138:
127:
122:
114:Find-me signals
110:
12:
11:
5:
451:
449:
441:
440:
435:
425:
424:
419:
418:
361:
297:
245:
181:
152:(4): 445–455.
124:
123:
121:
118:
117:
116:
109:
106:
25:Eat-me signals
13:
10:
9:
6:
4:
3:
2:
450:
439:
436:
434:
431:
430:
428:
414:
410:
405:
400:
396:
392:
388:
384:
381:(6): e52564.
380:
376:
372:
365:
362:
357:
353:
348:
343:
339:
335:
330:
325:
321:
317:
313:
306:
304:
302:
298:
293:
289:
285:
281:
277:
273:
269:
265:
261:
254:
252:
250:
246:
241:
237:
232:
227:
223:
219:
215:
211:
207:
203:
199:
192:
190:
188:
186:
182:
177:
173:
168:
163:
159:
155:
151:
147:
143:
136:
134:
132:
130:
126:
119:
115:
112:
111:
107:
105:
102:
98:
94:
90:
86:
82:
78:
77:glycoproteins
72:
70:
66:
62:
58:
54:
50:
46:
42:
41:phospholipids
38:
34:
30:
26:
18:
433:Cell biology
378:
375:EMBO Reports
374:
364:
319:
315:
267:
263:
205:
201:
149:
145:
89:phagocytosis
73:
57:calreticulin
24:
23:
208:(5): e331.
85:sialic acid
81:glycolipids
75:them. Most
69:pentraxin-3
39:, oxidized
33:phagocytose
427:Categories
322:: 629979.
120:References
61:annexin A1
29:phagocytes
395:1469-221X
338:1664-3224
292:221360052
222:1226-3613
49:galactose
413:34041845
356:34177884
284:32853880
240:28496201
176:22035837
146:Immunity
108:See also
101:opsonins
71:(PTX3).
65:histones
404:8183410
347:8220072
270:: 1–8.
231:5454446
167:3241945
97:mannose
55:(DNA),
411:
401:
393:
354:
344:
336:
290:
282:
238:
228:
220:
174:
164:
288:S2CID
45:sugar
409:PMID
391:ISSN
352:PMID
334:ISSN
280:PMID
236:PMID
218:ISSN
172:PMID
95:and
79:and
67:and
399:PMC
383:doi
342:PMC
324:doi
272:doi
226:PMC
210:doi
162:PMC
154:doi
51:),
31:to
429::
407:.
397:.
389:.
379:22
377:.
373:.
350:.
340:.
332:.
320:12
318:.
314:.
300:^
286:.
278:.
268:68
266:.
262:.
248:^
234:.
224:.
216:.
206:49
204:.
200:.
184:^
170:.
160:.
150:35
148:.
144:.
128:^
63:,
59:,
43:,
415:.
385::
358:.
326::
294:.
274::
242:.
212::
178:.
156::
Text is available under the Creative Commons Attribution-ShareAlike License. Additional terms may apply.